 |
 |
| Korean J Intern Med > Volume 38(4); 2023 > Article |
|
Abstract
Background/Aims
We evaluated the role of next-generation sequencing (NGS)-based disease monitoring for elderly patients diagnosed with acute myeloid leukemia (AML) who received decitabine therapy.
Methods
A total of 123 patients aged > 65 years with AML who received decitabine were eligible. We analyzed the dynamics of variant allele frequency (VAF) in 49 available follow-up samples after the fourth cycle of decitabine. The 58.6% VAF clearance (╬ö, [VAF at diagnosis ŌĆō VAF at follow-up] ├Ś 100 / VAF at diagnosis) was the optimal cut-off for predicting overall survival (OS).
Results
The overall response rate was 34.1% (eight patients with complete remission [CR], six of CR with incomplete hematologic recovery, 22 with partial responses, and six with morphologic leukemia-free status). Responders (n = 42) had significantly better OS compared with non-responders (n = 42) (median, 15.3 months vs. 6.5 months; p < 0.001). Of the 49 patients available for follow-up targeted NGS analysis, 44 had trackable gene mutations. The median OS of patients with ╬öVAF Ōēź 58.6% (n=24) was significantly better than that of patients with ╬öVAF < 58.6% (n = 19) (20.5 months vs. 9.8 months, p = 0.010). Moreover, responders with ╬öVAF Ōēź 58.6% (n = 20) had a significantly longer median OS compared with responders with VAF < 58.6% (n = 11) (22.5 months vs. 9.8 months, p = 0.004).
The incidence of acute myeloid leukemia (AML) increases with age, with a median age of approximately 70 years at diagnosis [1,2]. The prognosis of elderly patients with AML is poor for various reasons, including patientsŌĆÖ medical comorbidities, performance status, and disease biology [3ŌĆō5]. Compared with younger patients, elderly patients with AML have different genetic characteristics, and the treatment of elderly patients with AML remains challenging due to intolerance and resistance to intensive chemotherapy [4,6,7]. These factors lead physicians to favor less-intensive treatment rather than standard intensive chemotherapy [8,9]. Hypomethylating agents (HMAs) are widely used in clinical practice because they are relatively well tolerated with low treatment-related toxicity [10]. HMAs are particularly appropriate in the treatment of AML in elderly patients with comorbidities, poor performance status, and intolerance to combination therapy with venetoclax [10].
The genomic approach in clinical medicine has improved by developing next-generation sequencing (NGS). Thus, with NGS testing, AML can be categorized according to genetic risk [9,11]. However, the risk stratification of 2017 European LeukemiaNet (ELN) is generally focused on younger patients who are fit on standard induction treatment [11,12]. In addition, the therapeutic responses of AML patients were defined by morphologic complete remission (CR) [11]. Most patients with AML have molecular mutations at diagnosis, and NGS analysis has been widely used to detect and trace gene mutations. Therefore, NGS-based measurable residual disease (MRD) monitoring has been used to predict relapse in patients who have undergone intensive treatment [13,14]. Recent studies have been conducted to detect MRD to predict relapse risk [13ŌĆō17]. However, the genetic mutations associated with prognosis among older HMA-treated AML patients have not been well delineated, and an appropriate MRD marker for elderly AML patients has yet to be established [18,19].
In this retrospective study, we analyzed the prognostic impact of genetic mutations at AML diagnosis in elderly patients treated with decitabine. Additionally, we aimed to determine the prognostic significance of NGS-based disease monitoring in our cohort.
Patients diagnosed with AML between 2013 and 2020 were enrolled from a single institution. Eligible patients were those aged 65 years or older, with a confirmed diagnosis of AML according to the 2016 World Health Organization criteria, receiving decitabine for first-line therapy, and available for NGS using bone marrow (BM) samples at diagnosis. Patients diagnosed with acute promyelocytic leukemia were excluded. All patients received decitabine in standard doses (20 mg/m2 by intravenous infusion for 5 consecutive days) every 4 weeks. Among 64 patients who received at least four cycles of decitabine, 55 underwent follow-up BM biopsy after the fourth decitabine cycle, and two patients underwent BM biopsy after the second and third decitabine cycles, respectively, due to disease progression. Forty-nine patientsŌĆÖ follow-up BM samples underwent sequential targeted NGS sequencing. The study was conducted according to the principles of the Declaration of Helsinki. This study was approved by the Institutional Review Board of the Chonnam National University Hwasun Hospital, Korea (IRB No. CNUHH-2020-147). The details of the patients who underwent treatment are summarized in Supplementary Fig. 1.
NGS was performed on 123 samples collected from BM at the time of initial diagnosis and 49 follow-up BM samples. Deep sequencing was performed by targeting the coding regions of 51 genes with recurrent driver mutations based on data extracted from large cohort studies investigating AML and other myeloid malignancies (Supplementary Table 1) [20,21]. The targeted panel was constructed using a custom Agilent probe set (Agilent Technologies, Santa Clara, CA, USA). We defined the threshold of MRD positivity as variant allelic frequency (VAF) Ōēź 0.2% for follow-up BM biopsies based on the mutation locus background error rates, given that the mutations were present at diagnosis at VAF Ōēź 2% [13]. The detailed sample preparation, NGS, and variant calling procedures are provided in the Supplementary Materials.
Clinical characteristics were analyzed using chi-square tests for categorical variables, and two-sided StudentŌĆÖs t tests were used to analyze the quantitative variables. The statistical significance of VAF changes was determined using paired t tests. VAF clearance (╬öVAF) was calculated using the following formula: (VAFdx ŌĆō VAFfu) / VAFdx ├Ś 100. Using the R package ŌĆśrpartŌĆÖ for recursive partitioning to define a significant clearance rate (╬ö) of VAF, we defined ╬ö58.6% as the optimal cut-off value (Supplementary Fig. 2). For patients with multiple genetic alterations, the maximum VAF clearance rate was considered the ╬öVAF. The genetic risk stratification, along with definitions of CR, CR with incomplete hematologic recovery (CRi), morphologic leukemia-free state (MLFS), partial remission (PR), and no response, followed the 2017 ELN recommendations [11]. The overall response rate was defined as the proportion of patients who achieved CR, CRi, PR, and MLFS. Overall survival (OS) was calculated from the diagnosis of AML to the date of death or last follow-up. Event-free survival (EFS) was calculated as the interval from the date of the first administration of decitabine to the date of disease progression or date of death from any cause, whichever occurred first. The Kaplan-Meier method was used to analyze OS and EFS. The log-rank test was used to analyze the survival outcomes. Cox regression models were used for the multivariate analysis of various factors. To clarify the immortal time bias, landmark analyses were performed with patients (n = 84) who survived at least 3.6 months (because the median time from treatment initiation to follow-up BM examination after four decitabine cycles was 3.6 months). A p value less than 0.05 was considered statistically significant. Data analysis was performed using SPSS Statistics for Windows, version 26.0 (IBM Corp., Armonk, NY, USA) and EZR, version 1.54 (Jichi Medical University, Saitama, Japan) [22].
A total of 123 patients diagnosed with AML were eligible. With a median age of 75 years (range, 67 to 89 years), 112 patients (91.1%) were diagnosed with de novo AML and 11 (8.9%) with secondary AML. According to the 2017 ELN risk stratification, 22 patients (17.9%) were classified as favorable, 62 (50.4%) as intermediate, and 39 (31.7%) as adverse risk. Favorable cytogenetic abnormalities were detected in three patients (2.4%), and most patients (n = 71, 57.7%) had normal karyotypes. Twenty patients (16.3%) had complex aberrant karyotypes. The median number of decitabine cycles was 6 (range, 1 to 41), and the median follow-up duration was 15 months (range, 9.5 to 48.6 months) among survivors. There were 109 deaths (88.6%), and the most common causes of death were infection (43.1%) and disease progression (41.5%). The median OS was 6.3 months (95% confidence interval [CI], 5.1 to 8.1; Supplementary Fig. 3A), and the median EFS was 6.3 months (95% CI, 4.5 to 7.7; Supplementary Fig. 3B). Other details of the patientsŌĆÖ characteristics and clinical courses are summarized in Table 1 and Supplementary Fig. 1.
In the diagnostic samples, 108 patients (87.8%) had detectable genetic mutations, with a median of two mutations per patient (range, 1 to 6; mean, 2.3 per patient) and a median VAF of 32.5% (range, 2.0% to 95.0%). The patientsŌĆÖ mutational profile is shown in Supplementary Fig. 4. The most frequently detected mutations (in terms of categories of related genes) were those of activated signaling genes (n = 58; 47.2%), followed by DNA-methylationŌĆōrelated genes (n = 55, 44.7%), the gene encoding nucleophosmin 1 (n = 26, 21.1%), spliceosome-complex genes (n = 26, 21.1%), transcription-factor fusions (n = 21, 17.1%), and tumor-suppressor genes (n = 18, 14.6%). The most frequently detected mutations at diagnosis were mutations of FLT3 (n = 34, 27.6%; FLT3-ITD, n = 24 [high ratio, n = 10; low ratio, n = 14)]; FLT3-TKD, n = 10), followed by IDH1/IDH2 (24.3%; IDH1, n = 12; IDH2, n = 19), NRAS/KRAS (n = 18, 14.6%), DNMT3A (n = 18, 14.5%), TP53 (n = 15, 12.1%), and TET2 (n = 14, 11.0%) (Supplementary Fig. 4C). DNA methylation-related genes were more commonly detected in the intermediate ELN risk group than other risk groups (p = 0.019). Among 15 patients who carried no detectable mutations of known AML-associated driver genes, nine patients had normal karyotypes, and one patient had a complex karyotype.
Patients with complex karyotypes (n = 20; hazard ratio [HR], 1.52; 95% CI, 0.93 to 2.48; p = 0.095) showed a trend of poor survival compared with the other patients (Supplementary Fig. 5A). Patients with TET2 (n = 14; HR, 2.05; 95% CI, 1.15 to 3.63; p = 0.014) and TP53 (n = 15; HR, 1.64; 95% CI, 0.95 to 2.56; p = 0.097) mutations had poor OS. Patients with mutations of tumor-suppressor genes trended toward poor survival compared with the other groups (HR, 1.54; 95% CI, 0.92 to 2.56; p = 0.097). The survival analysis, conducted according to genetic mutations at diagnosis, is depicted in Supplementary Fig. 5B.
The overall response rate to decitabine treatment in this cohort was 34.1% (42/123; CR, n = 8; CRi, n = 6; PR, n = 22; MLFS, n = 6; Supplementary Fig. 6A). There were no significant differences in overall response rates according to 2017 ELN risk stratification, cytogenetic abnormality, genetic mutation, or category of related genes (Supplementary Fig. 6B, Table 2). Responders (n = 42; those who achieved CR, CRi, PR, or MLFS) had significantly better survival outcomes compared with non-responders (n = 42) in the land-mark analysis (median OS, 15.3 months vs. 6.5 months; p < 0.001; median EFS, 14.7 months vs. 6.5 months; p < 0.001) (Supplementary Fig. 3C, D). Among responders, the achievement of CR/CRi (n = 14) or PR (n = 22) was associated with better OS, but MLFS (n = 6) was associated with inferior OS (median OS, 19.3 months vs. 16.0 months vs. 8.1 months; p = 0.023) and inferior EFS (median EFS, 15.3 months vs. 15.1 months vs. 7.2 months; p = 0.036) (Supplementary Fig. 3E, F).
Of the 49 patients available for targeted NGS analysis after the fourth decitabine cycle, 44 had trackable gene mutations at diagnosis (Fig. 1). Eight of 44 patients (18.2%) achieved MRD negativity after four cycles of decitabine treatment. Among patients with each mutation, the median clearance rate of VAF was Δ43.7%. Figure 2 shows the clearance rates of VAF after the fourth decitabine cycle. Mutations of DNMT3A, TET2, and ASXL1 (DTA genes) were detected in samples from 29 patients at diagnosis. The DTA mutations, which are associated with age-related clonal hematopoiesis, were also persistently highly detected after the fourth cycle of decitabine (p = 0.428, Fig. 2B). Therefore, DTA mutations were excluded from the VAF clearance analysis. In the CR and CRi groups, the median VAFs at diagnosis decreased significantly after the fourth decitabine cycle (from 20.9% to 0.1% in the CR group, p = 0.001, Fig. 2C; from 20.6% to 2.9% in the CRi group, p = 0.121, Fig. 2D). In the PR group, the median VAF also decreased from 24.4% to 12.0% (p = 0.002, Fig. 2E). However, in the MLFS and non-responder groups, the median VAF was persistently maintained (from 18.7% to 19.9% in the MLFS group, p = 0.468, Fig. 2F; from 23.5% to 20.6% in the non-responder group; p = 0.665, Fig. 2G).
In the survival analysis, a 10% increase in mutation clearance improved OS by 11.7% (HR, 0.883; 95% CI, 0.837 to 0.930; p = 3.00 ├Ś 10ŌłÆ6). Based on a clearance rate of VAF ╬ö58.6%, which was the most informative cut-off point, 24 patients (54%) had VAF reductions Ōēź 58.6% (Fig. 1A). There were no associations of 2017 ELN risk stratification, cytogenetics, and categories of related gene abnormalities between responders and non-responders (Table 2). However, there was a significantly higher proportion of responders than non-responders (p = 0.043, Table 3) with reduced VAF (╬öVAF Ōēź 58.6%).
PTPN1, FLT3-TKD, EZH2, RUNX1, and TP53 mutations were associated with a median ╬öVAF Ōēź 58.6%. DNA-methylation gene (DMNT3A, TET2, IDH1, and IDH2) mutations were associated with a median ╬öVAF < 58.6% (Fig. 1B).
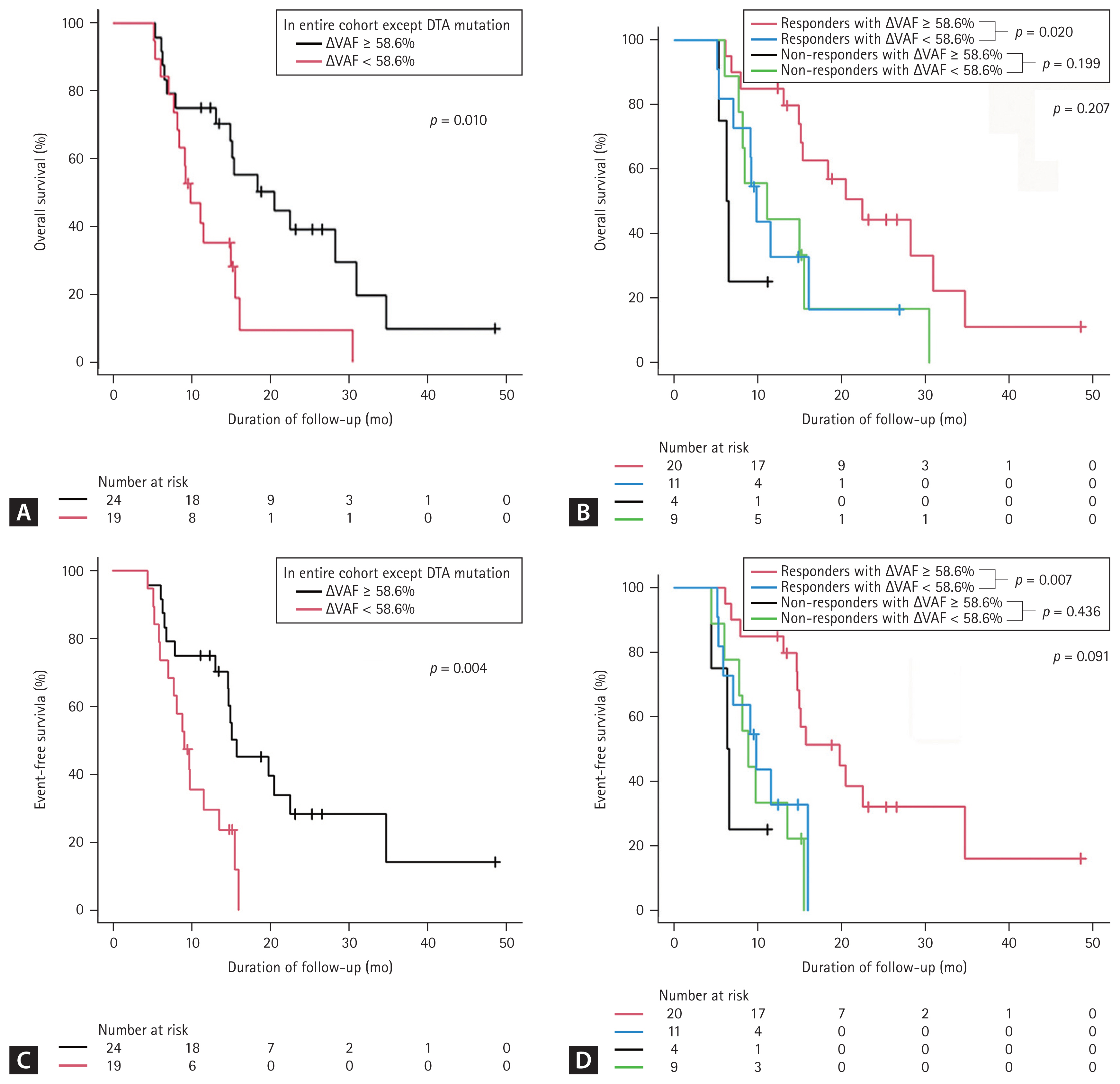
The median OS of patients with ╬öVAF Ōēź 58.6% (n = 24) was significantly longer than that of patients with ╬öVAF < 58.6% (n = 20 [median OS, 20.5 months vs. 9.8 months; p = 0.033]). MRD-negative patients had a good prognosis, with a median OS and EFS of 20.5 months, respectively. After DTA mutations were excluded, the survival outcomes remained similar to those derived from the analysis that included DTA mutations (median OS of ╬öVAF Ōēź 58.6% [n = 24], 20.5 months; ╬öVAF < 58.6% [n = 19], 9.8 months; p = 0.010; Fig. 3A). In addition, the median EFS of patients with reduced VAF was significantly longer than that of patients with ╬öVAF < 58.6% (median EFS, 15.7 months vs. 9.1 months, p = 0.007). In the analysis excluding the DTA mutations, the survival outcomes remained similar to those derived from the analysis that included the DTA mutations (median EFS of ╬öVAF Ōēź 58.6% [n = 24], 15.7 months; ╬öVAF < 58.6% [n = 19], 9.1 months; p = 0.004, Fig. 3C).
╬öVAF Ōēź 58.6% was more frequently observed in responders (p = 0.043, Table 3). Therefore, we further analyzed the prognostic significance of ╬öVAF Ōēź 58.6% in responders. Responders with ╬öVAF Ōēź 58.6% had significantly longer OS and EFS than responders with ╬öVAF < 58.6% (Fig. 3B, D). The median OS in the responders with ╬öVAF Ōēź 58.6% (n = 20) was 22.5 months, which was longer than the median 12.7 months in responders with ╬öVAF < 58.6% (n = 11, p = 0.020, Fig. 3B).
Table 4 summarizes the results of the univariate and multivariate Cox regression analyses to evaluate the variables associated with OS and EFS. Univariate analysis revealed ╬öVAF Ōēź 58.6%, TET2 mutation, and TP53 mutation to be significant predictors of OS and EFS (all, p < 0.05). Multivariate Cox regression analysis revealed ╬öVAF Ōēź 58.6% (OS [p = 0.047; HR, 0.46; 95% CI, 0.21 to 0.99], EFS [p = 0.019; HR, 0.39; 95% CI, 0.17 to 0.86]), TET2 (OS [p = 0.020; HR, 4.54; 95% CI, 1.27 to 16.25], and EFS [p = 0.019; HR, 4.67; 95% CI, 1.29 to 16.85]) to be independent predictors of OS and EFS (Table 4).
This study aimed to identify the prognostic significance of VAF reductions in elderly AML patients after decitabine treatment. The group with reduced VAF (╬öVAF Ōēź 58.6%) had significantly longer OS and EFS than the group without ╬öVAF Ōēź 58.6%. Additionally, responders with ╬öVAF Ōēź 58.6% had significantly longer OS and EFS. For elderly AML patients after decitabine treatment, long-term responses can be more accurately predicted when the ╬öVAF Ōēź 58.6%, with additional evidence from morphologic and hematologic HMA treatment responses.
MRD assessments are widely studied as prognostic factors after induction chemotherapy and after transplantation [13,14,23]. However, NGS-based MRD assessment in patients receiving low-intensity treatment has not been fully evaluated. The goal of intensive treatment is to achieve CR, and MRD clearance is an important predictable molecular marker of relapse. Recently, Boddu et al. [24] demonstrated the clinical relevance of MRD negativity in older patients with AML treated with HMAs. Using multicolor flow cytometry, they showed a significant advantage of MRD negativity in terms of the cumulative incidence of relapse [24]. However, this result did not translate to improved survival [24]. In that study, 13 patients were MRD-negative among 24 patients in the reduced-VAF group. The expectation of molecular clearance with only HMA treatment in older AML patients is questionable because somatic mutations with relatively high VAF remain after HMA administration [25,26]. Therefore, MRD negativity is not a conclusive prognostic marker for elderly AML patients treated with HMAs. In our study, reduced VAF (╬öVAF Ōēź 58.6%) was shown to be a feasible prognostic marker for predicting OS and EFS for elderly patients. These results emphasize the importance of VAF reduction, along with MRD negativity, in elderly AML patients treated with HMAs. The goal of HMA treatment in this patient population is to prolong survival rather than achieve cure.
Our study demonstrated that decitabine-treated elderly AML patients who achieved CR/CRi or PR had prolonged survival. Similar to our study, Molica et al. [27] found that the achievement of CR or PR after HMA treatment was significantly associated with better outcomes; they also observed no differences between CR and PR in elderly AML patients. However, that study did not clearly define MLFS. Our study found that such favorable outcomes were not associated with an MLFS in elderly AML patients. One of the reasons for the non-survival benefit in the MLFS group could be the accompanying infection and bleeding due to the cytopenia associated with an MLFS. Although limited in number, four out of six patients died from complications of cytopenia in our cohort. The other reason might be that an MLFS may make it difficult to predict a morphologic response due to the associated BM aplastic state. In our study, the MLFS patients had relatively stable VAFs compared with other responders (CR/CRi or PR)ŌĆöthe median ╬öVAFs were 11.5% vs. 88.0%, respectively. The MLFS can be interpreted as a depletion of leukemic cells but not a true decrease in leukemic burden compared with normal hematopoietic cells. Our study findings suggested that combining the MLFS evidence with ╬öVAF Ōēź 58.6% may address the limitations associated with the aplastic state in the MLFS.
Our data also demonstrate the prognostic significance of genetic mutations in the diagnosis of elderly AML patients receiving decitabine treatment. Metzeler et al. [28] found that the mutation spectrum of elderly AML patients differed from that of younger patients. The high rates of SRSF2, DNMT3A, TET2, and TP53 mutations in our elderly AML patients were concordant with other study findings [28,29]. These genes are age-associated and mediate clonal hematopoiesis. They are frequently mutated in elderly patients, and these results represent biological differences in the AML afflicting younger and elderly patients [30,31]. Unlike other studies, in our study, DNMT3A and RUNX1 mutations at diagnosis were not associated with inferior survival outcomes [32,33]. Differences in treatment modalities (intensive vs. non-intensive treatment) between studies could partially explain these conflicting findings. Since our study focused on the group receiving decitabine monotherapy, it was meaningful in that it demonstrated the significance of the genetic mutations in the HMA-treated group. Consistent with previous studies, we identified that TET2 and TP53 mutations at diagnosis were genetic risk predictors of poor survival [32,33]. It can be concluded that it is difficult to improve the survival of elderly AML patients with TP53 or TET2 mutations using HMA monotherapy. Elderly AML patients who carry these mutations could be recommended to register for clinical trials for early access to novel treatments. In our study, IDH1/IDH2 mutations were potentially associated with shorter survival in this patient population. A recent study found that patients with IDH1/IDH2-mutated AML had favorable outcomes when treated with HMAs and venetoclax [34]. Our data may provide additional evidence to recommend HMAs combined with venetoclax or IDH1/ IDH2 inhibitors because it is difficult to expect improved survival with HMAs alone in elderly AML patients with IDH1/ IDH2 mutations [35,36].
There were several limitations to our study. First, the use of ΔVAF is associated with selection bias because only patients who received at least four cycles of decitabine were included in the ΔVAF groups. For this reason, it is difficult to predict early mortality or early treatment failure at diagnosis. Second, this retrospective cohort included a small number of patients. However, our study provides appropriate evidence for applying NGS-based disease monitoring to elderly AML patients treated with HMAs. This study also suggests that a different approach to MRD monitoring is required for elderly AML patients.
Recently, HMAs with venetoclax has become a new standard treatment for older patients with AML [9,35,37]. However, the estimated median OS for patients with reduced VAF (╬öVAF Ōēź 58.6%) was 20.5 months, which was higher than that associated with the combination of HMA therapy and venetoclax (17.5 months) [38]. Some elderly AML patients are not suitable candidates for venetoclax plus HMAs because of treatment-related myelosuppression, comorbidities, and economic issues. ╬öVAF is an early predictor of long-term survival associated with decitabine monotherapy, and such prediction can help determine whether to continue treatment or consider other treatments at an early stage before the loss of the treatment response. Based on our data, VAF clearance could be an additional prognostic indicator of survival outcomes for elderly AML patients, particularly those who achieved PR or better responses after HMA treatment. Despite achieving overall responses to HMAs, patients with stable VAF may be unlikely to achieve long-term survival, and an early regimen change should be considered.
This study investigated the clinical significance of genetic mutations at the time of diagnosis in elderly AML patients receiving HMAs. Moreover, VAF clearance could provide additional information for predicting long-term survival in elderly AML patients responding to decitabine. ╬öVAF Ōēź 58.6% combined with evidence from morphologic and hematologic treatment responses could be proposed as a marker for determining whether to maintain decitabine treatment or combine novel agents to improve survival prognosis.
1. Variant allele frequency (VAF) clearance could provide additional information for predicting longterm survival in elderly acute myeloid leukemia patients responding to decitabine
2. VAF clearance with morphologic and hematologic responses could be proposed as a marker for determining whether to maintain decitabine treatment or combine novel agents to improve survival prognosis.
Acknowledgments
The biospecimens used in this study were provided by the Biobank of Chonnam National University Hwasun Hospital, a member of the Korea Biobank. The whole-exome data used in this study have been deposited in the Clinical & Omics Data Archive (CODA, http://coda.nih.go.kr) under accession # R000007.
Notes
CRedit authorship contributions
Mihee Kim: conceptualization, data curation, formal analysis, methodology, visualization, writing - original draft; TaeHyung Kim: data curation, methodology, visualization, writing - original draft; Seo-Yeon Ahn: data curation, formal analysis; Jun Hyung Lee: data curation, formal analysis; Ju Heon Park: methodology; Myung-Geun Shin: methodology; Sung-Hoon Jung: data curation, formal analysis; Ga-Young Song: data curation, formal analysis; Deok-Hwan Yang: formal analysis; Je-Jung Lee: formal analysis; Seung Hyun Choi: data curation, methodology; Mi Yeon Kim: data curation, methodology; Jae-Sook Ahn: conceptualization, formal analysis, funding acquisition, project administration, visualization, writing - review & editing; Hyeoung-Joon Kim: conceptualization, project administration; Dennis Dong Hwan Kim: conceptualization, project administration
Funding
This research was supported by the Basic Science Research Program, through the National Research Foundation of Korea (NRF), funded by the Ministry of Science, ICT, and Future Planning (NRF-2015R1A2A1A10054579) and the National R&D Program for Cancer Control, Ministry of Health & Welfare, Republic of Korea (1720160). This study was supported by a grant (HCRI21006) from Chonnam National University Hwasun Hospital Institute for Biomedical Science. This work was also supported by a National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (No. 2018R1A2A1A05078480, 2022R1F1A1063836) and by Foundation of cutting-edge supporting infrastructure for precision medicine industry (P0017144).
Figure┬Ā1
Variant allele frequency clearance rate (ΔVAF) at follow-up; ΔVAFs measured from paired samples at diagnosis and the follow-up bone marrow samples after four cycles of decitabine. (A) ΔVAFs at follow-up, (B) ΔVAF at follow-up in association with each genetic mutation.
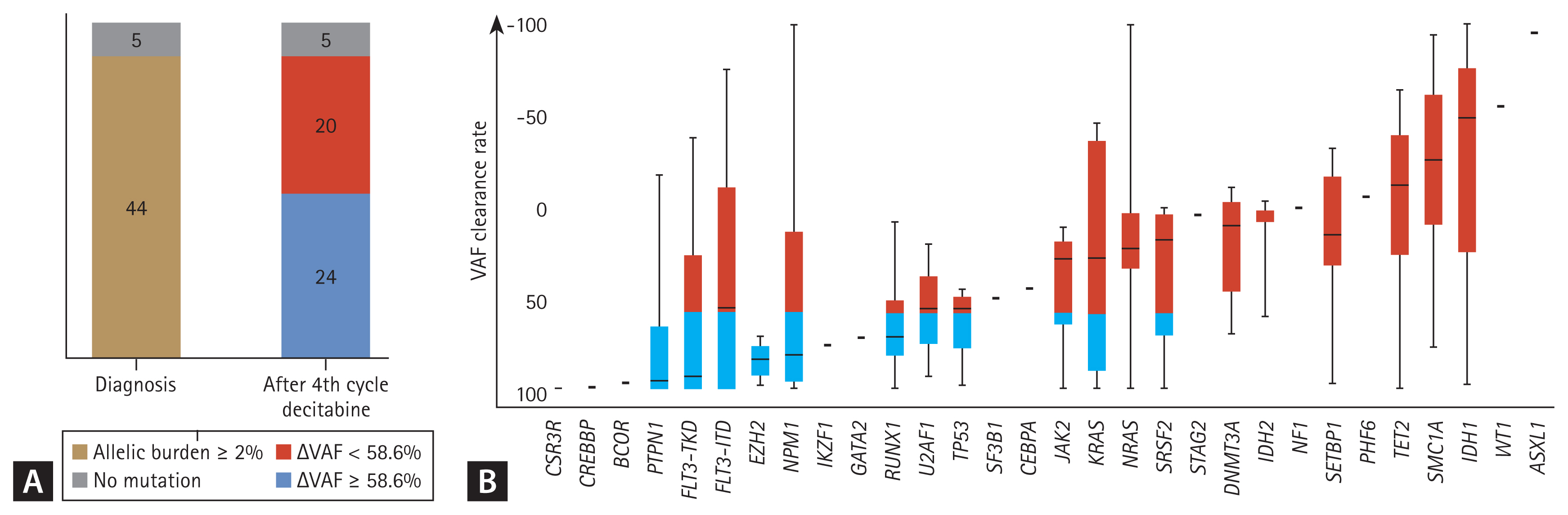
Figure┬Ā2
Dynamics of variant allele frequencies (VAFs) of mutations at two time points (at diagnosis and after the fourth cycle of decitabine) in elderly acute myeloid leukemia HR patients. (A) Entire cohort; (B) group with DNMT3A, TET2, and ASXL1 (DTA) mutations; (C) complete remission (CR) group; (D) complete remission with incomplete hematologic recovery (CRi) group; (E) partial remission (PR) group; (F) morphological leukemia-free state (MLFS) group; and (G) non-responders.

Figure┬Ā3
Kaplan-Meier curves using landmark analysis (3.6 months) for elderly acute myeloid leukemia patients according to variant allele frequency VAF clearance (ΔVAF). (A) Overall survival (OS) in the entire cohort excluding those with DNMT3A, TET2, and ASXL1 (DTA) mutations; (B) OS in the non complete remission (non-CR)/CR with incomplete hematologic recovery (CRi) group; (C) event-free survival (EFS) in the entire cohort excluding those with DTA mutations; (D) EFS in the non-CR/CRi group. After landmark analysis, the 39 patients who died before 3.6 months were excluded from the survival analysis. Others included the stable VAF group (ΔVAF < 58.6%), the group of patients with no mutations, and the group that did not have available follow-up BM samples in each cohort.
Table┬Ā1
Clinical characteristics of AML patients (n = 123)
Table┬Ā2
Clinical correlation of overall response after four cycles of decitabine (n = 64)
Table┬Ā3
Clinical correlation between ╬öVAF Ōēź 58.6% and ╬öVAF < 58.6% after the fourth cycle chemotherapy (n = 44)a)
Table┬Ā4
Univariate and multivariate Cox proportional hazards regression analyses predicting OS and EFS in elderly AML patients
| Variable | Overall survival | Event free survival | ||||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||
| Univariate HR (95% CI) | p value | Multivariate HRa) (95% CI) | p value | Univariate HR (95% CI) | p value | Multivariate HRa) (95% CI) | p value | |
| Ageb) | 1.06 (1.01ŌĆō1.11) | 0.009 | 1.09 (0.97ŌĆō1.23) | 0.149 | 1.05 (1.01ŌĆō1.10) | 0.020 | 1.06 (0.95ŌĆō1.19) | 0.309 |
|
|
||||||||
| TET2 | 2.05 (1.15ŌĆō3.63) | 0.014 | 4.54 (1.27ŌĆō16.25) | 0.020 | 2.00 (1.13ŌĆō3.55) | 0.018 | 4.67 (1.29ŌĆō16.85) | 0.019 |
|
|
||||||||
| TP53 | 1.65 (0.95ŌĆō2.86) | 0.077 | 3.97 (1.12ŌĆō14.04) | 0.032 | 1.77 (1.02ŌĆō3.07) | 0.044 | 1.72 (0.32ŌĆō9.29) | 0.530 |
|
|
||||||||
| Complex karyotype | 1.52 (0.93ŌĆō2.48) | 0.095 | 0.98 (0.26ŌĆō3.69) | 0.978 | 1.76 (1.07ŌĆō2.89) | 0.026 | 3.79 (1.43ŌĆō10.07) | 0.008 |
|
|
||||||||
| ΔVAFc) | ||||||||
|
|
||||||||
| Ōēź 58.6% | 0.47 (0.22ŌĆō0.98) | 0.049 | 0.46 (0.21ŌĆō0.99) | 0.047 | 0.36 (0.17ŌĆō0.80) | 0.012 | 0.39 (0.17ŌĆō0.86) | 0.019 |
|
|
||||||||
| < 58.6% | Reference | Reference | ||||||
Univariate analysis and multivariate Cox proportional hazards regression analysis was conducted using the log-rank method. Multivariate analysis was performed using the covariates that yielded p values < 0.1 in the univariate analysis.
OS, overall survival; EFS, event-free survival; AML, acute myeloid leukemia; HR, hazard ratio; CI, confidence interval; VAF, variant allele frequency; ΔVAF, VAF clearance rate.
REFERENCES
1. Prassek VV, Rothenberg-Thurley M, Sauerland MC, et al. Genetics of acute myeloid leukemia in the elderly: mutation spectrum and clinical impact in intensively treated patients aged 75 years or older. Haematologica 2018;103:1853ŌĆō1861.



2. Thein MS, Ershler WB, Jemal A, Yates JW, Baer MR. Outcome of older patients with acute myeloid leukemia: an analysis of SEER data over 3 decades. Cancer 2013;119:2720ŌĆō2727.



3. Hourigan CS, Karp JE. Development of therapeutic agents for older patients with acute myelogenous leukemia. Curr Opin Investig Drugs 2010;11:669ŌĆō677.


4. Ossenkoppele G, L├Čwenberg B. How I treat the older patient with acute myeloid leukemia. Blood 2015;125:767ŌĆō774.



6. Klepin HD. Elderly acute myeloid leukemia: assessing risk. Curr Hematol Malig Rep 2015;10:118ŌĆō125.




7. Rao AV, Valk PJ, Metzeler KH, et al. Age-specific differences in oncogenic pathway dysregulation and anthracycline sensitivity in patients with acute myeloid leukemia. J Clin Oncol 2009;27:5580ŌĆō5586.

8. Rao AV. Fitness in the elderly: how to make decisions regarding acute myeloid leukemia induction. Hematology 2016;2016:339ŌĆō347.




10. Dombret H, Seymour JF, Butrym A, et al. International phase 3 study of azacitidine vs conventional care regimens in older patients with newly diagnosed AML with >30% blasts. Blood 2015;126:291ŌĆō299.




11. D├Čhner H, Estey E, Grimwade D, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood 2017;129:424ŌĆō447.




12. Wang B, Liu Y, Hou G, et al. Mutational spectrum and risk stratification of intermediate-risk acute myeloid leukemia patients based on next-generation sequencing. Oncotarget 2016;7:32065ŌĆō32078.



13. Ahn JS, Kim T, Jung SH, et al. Allogeneic transplant can abrogate the risk of relapse in the patients of first remission acute myeloid leukemia with detectable measurable residual disease by next-generation sequencing. Bone Marrow Transplant 2021;56:1159ŌĆō1170.



14. Kim T, Moon JH, Ahn JS, et al. RNA sequencing as an alternative tool for detecting measurable residual disease in core-binding factor acute myeloid leukemia. Sci Rep 2020;10:20119.




15. Thol F, Gabdoulline R, Liebich A, et al. Measurable residual disease monitoring by NGS before allogeneic hematopoietic cell transplantation in AML. Blood 2018;132:1703ŌĆō1713.




16. Ivey A, Hills RK, Simpson MA, et al. Assessment of minimal residual disease in standard-risk AML. N Engl J Med 2016;374:422ŌĆō433.


17. Thol F, K├Člking B, Damm F, et al. Next-generation sequencing for minimal residual disease monitoring in acute myeloid leukemia patients with FLT3-ITD or NPM1 mutations. Genes Chromosomes Cancer 2012;51:689ŌĆō695.


18. Krug U, Gale RP, Berdel WE, et al. Therapy of older persons with acute myeloid leukaemia. Leuk Res 2017;60:1ŌĆō10.


19. Hilberink JR, Morsink LM, van der Velden WJFM, et al. Pre-transplantation MRD in older patients with AML after treatment with decitabine or conventional chemotherapy. Transplant Cell Ther 2021;27:246ŌĆō252.


20. Papaemmanuil E, Gerstung M, Bullinger L, et al. Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med 2016;374:2209ŌĆō2221.



21. Ley TJ, Miller C, Ding L, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med 2013;368:2059ŌĆō2074.



22. Kanda Y. Investigation of the freely available easy-to-use software ŌĆśEZRŌĆÖ for medical statistics. Bone Marrow Transplant 2013;48:452ŌĆō458.




23. Jongen-Lavrencic M, Grob T, Hanekamp D, et al. Molecular minimal residual disease in acute myeloid leukemia. N Engl J Med 2018;378:1189ŌĆō1199.


24. Boddu P, Jorgensen J, Kantarjian H, et al. Achievement of a negative minimal residual disease state after hypomethylating agent therapy in older patients with AML reduces the risk of relapse. Leukemia 2018;32:241ŌĆō244.



25. Falconi G, Fabiani E, Piciocchi A, et al. Somatic mutations as markers of outcome after azacitidine and allogeneic stem cell transplantation in higher-risk myelodysplastic syndromes. Leukemia 2019;33:785ŌĆō790.




26. Buccisano F, Dillon R, Freeman SD, Venditti A. Role of minimal (measurable) residual disease assessment in older patients with acute myeloid leukemia. Cancers (Basel) 2018;10:215.



27. Molica M, Mazzone C, Niscola P, et al. Identification of predictive factors for overall survival and response during hypomethylating treatment in very elderly (Ōēź75 years) acute myeloid leukemia patients: a multicenter real-life experience. Cancers (Basel) 2022;14:4897.



28. Metzeler KH, Herold T, Rothenberg-Thurley M, et al. Spectrum and prognostic relevance of driver gene mutations in acute myeloid leukemia. Blood 2016;128:686ŌĆō698.



29. Tsai CH, Hou HA, Tang JL, et al. Genetic alterations and their clinical implications in older patients with acute myeloid leukemia. Leukemia 2016;30:1485ŌĆō1492.



30. Genovese G, K├żhler AK, Handsaker RE, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med 2014;371:2477ŌĆō2487.



31. Park DJ, Kwon A, Cho BS, et al. Characteristics of DNMT3A mutations in acute myeloid leukemia. Blood Res 2020;55:17ŌĆō26.




32. Tsai CH, Hou HA, Tang JL, et al. Genetic alterations and their clinical implications in older patients with acute myeloid leukemia. Leukemia 2016;30:1485ŌĆō1492.



33. Wang SY, Cheng WY, Mao YF, et al. Genetic alteration patterns and clinical outcomes of elderly and secondary acute myeloid leukemia. Hematol Oncol 2019;37:456ŌĆō463.




34. Pollyea DA, Dinardo CD, Arellano ML, et al. Results of venetoclax and azacitidine combination in chemotherapy ineligible untreated patients with acute myeloid leukemia with IDH 1/2 mutations. Blood 2020;136:5ŌĆō7.


35. Park S, Cho BS, Kim HJ. New agents in acute myeloid leukemia (AML). Blood Res 2020;55:S14ŌĆōS18.



36. Byun JM, Yoo SJ, Kim HJ, et al. IDH1/2 mutations in acute myeloid leukemia. Blood Res 2022;57:13ŌĆō19.



- TOOLS
-
METRICS

-
- 0 Crossref
- 0 Scopus
- 256 View
- 122 Download
- Related articles
-
Mixed-phenotype acute leukemia treated with decitabine2016 March;31(2)



 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Download Citation
Download Citation Supplement figure 1
Supplement figure 1 Print
Print


