 |
 |
| Korean J Intern Med > Volume 18(1); 2003 > Article |
|
Abstract
Background
The t (8;21) (q22;q22), which produces the fusion gene AML1/ETO, is associated with relatively good prognosis and, in particular, with a good response to cytosine arabinoside. Analysis of t (8;21) positive leukemic blasts has shown characteristic morphological and immunological features. We performed this study to investigate the incidence of AML1/ETO rearrangement in adult acute myelogenous leukemia (AML), especially in M2 subtype, to make a comparison of clinical, morphological and immunophenotypic characteristics between AML1/ETO rearrangement positive and negative group in patients with AML and to analyze the correlation with other biological parameters.
Methods
From May 1995 to Sept. 2000, fifty-nine patients with AML, including twenty-nine AML-M2, were studied. RNAs were extracted from leukemic cells and reverse transcriptase mediated polymerase chain reaction (RT-PCR) for AML1/ETO fusion transcript was done. Chromosome study, immunophenotypic and clinical characteristics were analyzed and statistical analysis was done.
Results
The incidence of AML1/ETO fusion transcripts was 22.0% in AML and 44.8% in AML-M2. The morphologic finding of bone marrow in AML-M2 showed higher incidence of Auer rods, large blast with prominent golgi and abnormal granules in AML1/ETO positive patients. There was no significant difference of immunophenotype.
AML patients with AML1/ETO had a tendency of higher complete remission rate (81.8% vs 56.6%, p=0.13). The overall survival (median; 82.2 weeks vs 34.4 weeks, p=0.02) and progression free survival (median; 50.9 weeks vs 20.4 weeks, p=0.02) of AML1/ETO positive group were longer than those of the negative group in AML. AML-M2 patients with AML1/ETO rearrangement had also a tendency of longer overall survival and progression free survival, although there was no significant difference between both groups.
Morphological and cytogenetic characteristics in acute myelogenous leukemia (AML) are the most important prognostic factors and are used in diagnosis and treatment, as well as in understanding their pathophysiology. Chromosomal abnormality, known as relatively good prognostic factors, are t (8;21) (q22;q22), t (15;17) (q22;q21) and inv (16), of which t (8;21) (q22;q22) is most frequent in AML and has specific clinical, morphological and immunological characteristics.
The incidence of t (8;21) (q22;q22) is higher in younger patients and it is often associated with extramedullary tumors or chloromas and splenomegaly1, 2). Morphologically typical t (8;21) positive leukemic blasts are large with abundant cytoplasm, numerous granules and occasional homogenous salmon-coloured granules. Prominent Auer rods and cytoplasmic vacuoles are frequently seen. These are associated with smaller immature blasts with frequent eosinophilia in the bone marrow2ŌĆō4). In addition, the t (8;21) positive leukemic blasts frequently have a specific immunophenotype5, 6), characterized by positivity for the B-cell-associated marker CD19, as well as CD13, CD34, CD56 and HLA-DR. In contrast, CD2 and CD7 are rarely expressed and CD33 is characteristically weak. The presence of t (8;21) is associated with a high remission rate and prolonged disease-free survival in patients treated with cytosine arabinoside in chemotherapy7), but is of no advantage to high-dose chemotherapy and marrow transplantation8). Over 90% of t (8;21) is found in AML-M2, 6% in M1 and rare in another group like M49).
From the view of molecular genetics, gene translocation is occurred between the AML1 gene on chromosome 21q22 and the ETO gene on chromosome 8q22 to form new AML1/ETO fusion gene, concerning pathophysiology of leukemia10, 11). In spite of the prognostic importance of detection of the t (8;21) in AML, most institutes were not able to perform adequate cytogenetic analysis in a large number of AML patients. However, it is possible to detect this translocation by molecular techniques, including fluorescence in situ hybridization (FISH), Southern blotting and, most commonly, reverse transcriptase mediated polymerase chain reaction (RT-PCR).
Kim et al. reported the morphological and immunophenotypic characteristics of myeloid malignancies with t (8;21)12) and Lee et al. reported AML1/ETO fusion gene expression and their clinical features in AML13), but they were small in number and there was no report about the relationship with the prognosis. In this study, we investigated the frequency of AML1/ETO fusion transcripts in AML, accordingly, any differences in hematological and clinical features and the relationship with the prognosis. Also, the frequency of AML1/ETO fusion transcripts in AML-M2 patients and their relationship with the prognosis were reviewed.
Patients diagnosed as AML at Gachon Medical School, Gil Medical Center from May 1995 to Sept. 2000 were studied, except M3 patients with t (15;17). Morphological diagnosis and classification were based on analysis of peripheral blood and bone marrow smears stained by May-Gr├╝nwald-Giemsa and on cytochemical staining for myeloperoxidase, according to the French-American-British (FAB) cooperative criteria. Particular attention was paid to the presence of morphological features considered to be suggestive of a t (8;21). Clinical and hematologic data and characteristics of patients were analyzed and used for statistical analysis.
Bone marrow aspirate treated by heparin was put in a 25 cm2 flask of non-PHA culture medium (RPMI 1640, 20% fetal bovine serum, penicillin-streptomycin) and cultured at 37┬░C and 5% CO2 for 24 hours. After culturing with Colcemid, it was treated with 0.075M KCI hypotonic solution and fixed with a fixative (glacial acetic acid : methanol 1:3) over 3 times. Cell suspension was dropped on the slide before drying and aging for 2 days. The trypsin-Giemsa method was used for G- banding. Karyotyping was performed according to the International System for Human Cytogenetic Nomenclature (ISCN 1995).
Bone marrow aspirates were collected into a preservative free heparin tube (Gibco, Grand Island, NY, USA). Mononuclear cells obtained by Ficoll-Hypaque (Pharmacia, Picataway, NJ, USA) density gradient centrifugation were made to 0.5ŌĆō2├Ś107/mL of concentration before RNA extraction or storage at ŌłÆ70┬░C.
Total RNA was extracted from leukemic cells by the guanidium thiocyanate-phenol-chloroform method. cDNA was synthesized from 1 ╬╝g of RNA with 100 U of MoMLV reverse transcriptase (Promega, USA) in 1├ŚPCR reaction buffer containing 100 pmol of random hexamer primers, 10 mM dNTP, 20 U of RNase inhibitor (Promega, USA) in a total volume of 20 ╬╝L at 37┬░C for 1 hour. In order to inactivate reverse transcriptase, the reacted product was treated at 94┬░C for 5 minutes. cDNA synthesis was confirmed by using ╬▓-actin primer to perform PCR and then electrophoresed in a 1% agarose gel with ethidium bromide staining and observed by polaroid photos.
The first PCR amplification was accomplished in a volume of 100 ╬╝L using 10 ╬╝L of the cDNA mixture additional 1 ├Ś PCR reaction buffer containing 1.25 U Taq polymerase (Promega, USA), 10 pmol/L of oligonucleotide primers14) (Table 1). The PCR amplification was performed on a DNA thermal cycler (Ericomp, USA) with an initial denaturation step of 94┬░C for 5 minutes followed by 35 cycles of denaturation at 94┬░C for 1 minute, annealing at 55┬░C for 1 minute, and extension at 72┬░C for 1 minute. A final elongation step at 72┬░C for 10 minutes was done. The second PCR amplification was performed under identical cycling conditions using 1 ╬╝L of first PCR products and a set of nested primers.
The nested PCR products were separated by electrophoresis in a 1% agarose gel with ethidium bromide staining and observed by polaroid photos.
Bone marrow aspirate 100 μL was added with and stained directly by 20 μL of CD3, CD5, CD7, CD10, CD19, CD20, CD22, CD14, CD13, CD33, CD45 and HLA-DR monoclonal antibody (Immunotech, A Coulter Company, 13009 Marseille, France) each and analyzed by flow cytometry Coulter EPIX® XL (Coulter Corporation, Miami, USA).
Induction chemotherapy used cytarabine and anthracycline. Cytarabine 100 mg/m2/day was continuously intravenous-infused from 1st day to 7th day, and idarubicine/mitoxantrone 12 mg/m2/day or daunorubicin 40 mg/m2/day was infused from 1st day to 3rd day. Also, in case of adding etoposide to cytarabine and idarubicine, 75 mg/m2/day was infused from 1st day to 5th day. Marrow examination was performed on 28th day after remission induction therapy, and it was determined as a complete response when the leukemic blasts count within the marrow was less than 5%, and neutrophils>1,000/╬╝L and platelets> 100,000/╬╝L in peripheral blood were continued over 4 weeks.
Relapse-free survival duration was calculated in patients who obtained a complete remission after induction chemotherapy from the day determined as a complete response to the day of relapse or death. Progression-free survival was measured in all the patients from the day of diagnosis to the day of relapse or death. The survival duration was from the day of diagnosis to the day of death or last follow-up for all the patients. Any difference of the absolute value in each group was compared by Nonparametric test (Mann-Whitney test), and ratio difference by Fisher's exact test. Survival curve was calculated by the Kaplan-Meier method and its comparison adopted the log-rank test.
A total of 59 patients were studied, of whom 4 patients showed secondary AML converted from myelodysplastic syndrome. The gender ratio was 32:27 (M:F), and the median age was 43 years (ranging 14ŌĆō86). The frequency by FAB classification standard appeared as M0;5, M1;13, M2;29, M4;3, M5;2, M6;4, M7;3 patients. AML1/ETO transcripts were detected at 338 bp in the first PCR products and at 185 bp in the second PCR products, which was found in 22.0% (13/59) and ╬▓-actin was all positive (Figure 1). 36 patients underwent the cytogenetic test and t (8;21) (q22;q22) was expressed by 27.8% (10/36).
AML-M2 patients were 29, whose gender ratio was 17:12 (M:F) and the median age was 43 years (ranging 14ŌĆō77). AML1/ETO transcripts in M2 were detected in 44.8% (13/29). After the cytogenetic test with 20 patients, t (8;21) (q22;q22) expression was 50.0% (10/20). Only one patient had AML1/ETO transcripts without t (8;21).
In this study, we made a comparison of the clinical parameters such as age, gender, hepatomegaly, splenomegaly and extramedullary tumefaction between AML1/ETO positive and negative group in AML patients, but did not find a significant difference. Only the mean value of leukocyte was 50.0├Ś103/╬╝L in the negative group and 19.2├Ś103/╬╝L in the positive group, which was higher in the negative group. The median of blast % within the bone marrow was 58% in the positive group and 80% in the negative group, which was higher in the negative group, but of no significance (Table 2). These factors were compared in M2 patients, but there was no significant difference.
33 patients in AML underwent immunophenotyping and a higher frequent expression of HLA-DR was found in the positive group, but it was not statistically significant (p=0.06) (Table 3). 15 patients in AML-M2 underwent immunophenotyping, which was not significantly different.
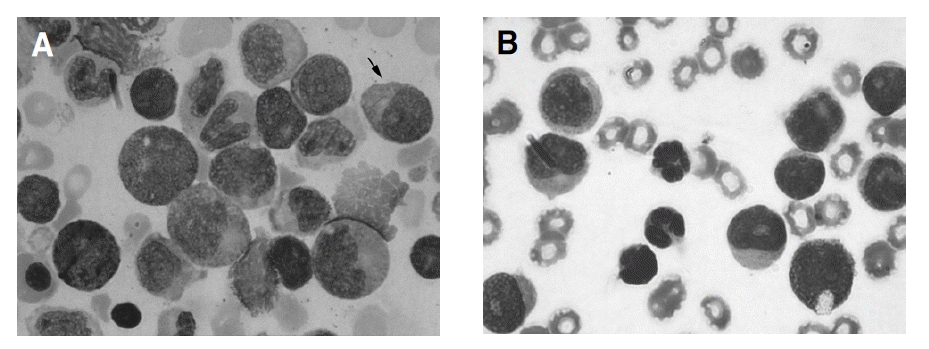
As a result of analysis on the morphologic characteristics of 29 AML-M2 patients (Table 4), Auer rods in leukemic blasts were more observed and lots of large blasts with prominent golgi and abnormal granules were found more in AML1/ETO positive patients (Figure 2). The positive patients showed eosinophilia over 3% of all nucleated cell within the bone marrow, which was not statistically significant.
41 patients underwent chemotherapy, in which cytarabine + idarubicine were for 29 patients, cytarabine + mitoxantrone/daunorubicin for 6 and cytarabine + idarubicine + etoposide for 5. Complete remission rate of the overall chemotherapy group was 63.4% (26/41), AML1/ETO positive group showed 81.8% (9/11), while the negative group showed 56.6% (17/30), which was of no statistical significance, but the complete remission rate was higher in the positive group (p=0.13). The median relapse-free survival rates in patients with complete remission were 44.0 weeks (95% confidence interval [CI]: 35.5~52.5) for patients with AML, 46.3 weeks (95% Cl: 5.4~87.2) for AML1/ETO positive group, 42.7 weeks (95% Cl: 31.4~54) for AML1/ETO negative group, and there was no statistical significance between the two groups (p=0.24). The median survival duration during 109 weeks of median follow-up displayed 40.3 weeks (95% Cl: 22.6~58.0) for AML patients, 82.2 weeks (95% Cl: 10.5~154.3) for AML1/ETO positive group, 34.4 weeks (95% Cl: 7.1~61.7) for AML1/ETO negative group, by which it was significantly longer in the positive group (p=0.02) (Figure 3). The median progression-free survival rates were 25.0 weeks (95% Cl: 9.4~40.6) for AML patients, 50.9 weeks (95% Cl: 8.1~93.7) for AML1/ETO positive group, 20.4 weeks (95% Cl: 8.1~32.7) for AML1/ETO negative group, which was significantly longer in the positive group (p=0.02) (Figure 4).
20 patients in AML-M2 underwent chemotherapy and the complete remission was 70% (14/20). The complete remission rates in AML1/ETO positive and negative were 81.8% (9/11) and 55.5% (5/9), which were not statistically significant, but higher in the positive group. The median relapse-free survival rates in patients with complete remission were 46.3 weeks (95% Cl: 5.4~87.2) for AML1/ETO positive group and 76.6 weeks (95% Cl: 0.2~168.9) for AML1/ETO negative group, which were not significantly different. The medial follow-up duration of M2 patients was 97 weeks and the median survival duration was 50.4 weeks (95% Cl: 0.2~100.6), in which it was 82.4 weeks (95% Cl: 10.5~154.3) for AML1/ETO positive group and 15.6 weeks (95% Cl: 0.2~39.6) for AML1/ETO negative group. It was not statistically significant, but it was higher in the positive group (p=0.07). The median progression-free survival rates were 50.9 weeks (95% Cl: 8.1~93.7) in the positive group and 16.0 weeks (95% Cl: 0.2~34.1) in the negative group, which was not statistically significant, but longer in the positive group.
The t (8;21) (q22;q22) is a good prognostic factor of AML and found in 7~10% of AML and 20% of AML-M2. The frequency of AML1/ETO fusion transcripts by RT-PCR is found in 13% of AML and 30ŌĆō40% of AML-M215). In this study, t (8;21) (q22;q22) was observed in 27.8% of AML and high 50% of AML-M2. The frequency of AML1/ETO fusion transcripts by RT-PCR was positive in 22.0% of AML and 44.8% of AML-M2. As this is a bit higher frequency than other reports, it requires a further follow-up study with more patients.
It is usual that chromosome karyotyping and molecular genetic lesion by RT-PCR are not completely coincident. The incidence of AML1/ETO might increase by 5ŌĆō10% by the molecular biological approach, like RT-PCR, which had demonstrated the occasional presence of this molecular lesion in cases that lack t (8;21)16, 17). This finding suggests that molecular diagnostic approaches should be routinely used to identify patients with this genetic subtype of AML. It was reported to be a poor prognosis in AML1/ETO positive cases by RT-PCR without t (8;21)18). This study found one AML1/ETO positive patient without t (8;21), whose relapse-free survival was 8 weeks and overall survival was 16 weeks. Of patients who didnot get the result by chromosome testing in this study, AML1/ETO positive patients were 2 cases. Zhang et al. reported that patients with t (8;21) showed AML1/ETO negative by Southern and/or PCR analysis because the breakpoint within AML1 was located downstream to the breakpoint cluster19). Therefore, it is clear that such a sensitive and rapid molecular biological diagnosis as RT-PCR and chromosome testing should be basically used in determining acute myelogenous leukemia subtypes.
The morphologic features of t (8;21) leukemic blasts are large with abundant cytoplasm, lots of granules and occasional pseudo-Chediak granules. Vacuoles and Auer rods in cytoplasm are frequently seen2ŌĆō4). Kim et al.12) observed Auer rods in 67% (14/21) of AML-M2 and myelodysplastic syndrome with t (8;21). As a result of a comparison between AML1/ETO positive and negative patients group in AML M2 for morphologic features in this study, cells with Auer rods and lots of granules were more frequently observed in the positive group than in the negative group. Also, large blasts with prominent golgi were significantly more observed in the positive group. There was a tendency of lower blasts count and eosinophilia within the bone marrow in the positive group, which was not statistically significant.
The t (8;21) leukemic blasts frequently have a distict immunophenotype, characterized by positivity for the B-cell-associated marker CD19, as well as CD13, CD34 and CD56, and high HLA-DR expression, while poor CD33 expression6, 7). Kim et al. reported that atypical CD19 was significantly high expressed by 83%, but CD33 was expressed significantly lower than other markers12). In this study, only 1 of 8 AML1/ETO positive patients examined by immunophenotyping expressed CD19 and 5 patients expressed CD33 and, accordingly, any statistical significance was not found. HLA-DR expression in AML1/ETO positive group was high compared to the negative group (p=0.06).
Cytarabine, a major source in induction chemotherapy of AML, shows a complete response in 50~80% of patients under combination with anthracycline. It is known that a complete remission rate is high and remission duration or survival rate is significantly high also in AML with t (8;21)20). Relapse-free survival rate is known to be increased after intensification therapy with high dose cytarabine8). More recently, it has been demonstrated that treatment of this group of patients with chemotherapy alone results in cure rates that are comparable to or better than those achieved with allogeneic bone marrow transplantation21). Therefore, identification of these molecular genetic lesions at the time of diagnosis may spare patients from the risks associated with allogeneic bone marrow transplantation. As a result of this study, the remission rate of AML1/ETO positive group with AML was higher than the negative group, which was of no statistical significance, and there was no difference in the relapse-free survival rate. But both overall survival and progression-free survival rates were significantly higher in the positive group. The remission rate in AML-M2 was higher in the positive group, which was not statistically significant, and the relapse-free survival was not different. Overall survival and progression-free survival rates were higher in the positive group, which were not statistically significant.
The application of these molecular approaches to monitor patients for evidence of so-called minimal residual disease (MRD) has been controversial. Several studies have demonstrated persistent expression of the AML1/ETO chimeric mRNA in patients in long-term remission, including those treated with allogeneic bone marrow transplantation22, 23). In addition, persistent expression of AML1/ETO was detected in the clonogenic hematopoietic progenitors isolated from remission bone marraw samples obtained from patients who had been in remission off therapy for several years. AML1/ETO expression was detected not only in myeloid/monocytic colonies, but also in erythroid colonies, suggesting that despite being in remission the bone marrow of these patients had retained viable multipotential hematopoietic progenitors containing t (8;21). More recently, several studies have failed to detect expression of AML1/ETO in the bone marrows of patients in long-term remission24ŌĆō26). In prospectively studied patients, a good correlation was found between negative PCR results and absence of relapse24). Early negative results with the one-step RT-PCR technique, before consolidation treatment, seemed to carry an especially good prognosis, suggesting that RT-PCR analysis could help in choosing the type of consolidation therapy in patients with t (8;21). This study performed a follow-up AML1/ETO molecular study on four AML1/ETO positive patients in complete remission after treatment. 2 of them experienced relapse of leukemia in a persistent AML1/ETO expression, while 2 patients were converted into the negative group and maintained in remission without recurrence for 106 weeks and 71 weeks, respectively. The recent development of quantitative RT-PCR assays for chimeric AML1/ETO transcripts should in the future provide a valuable tool for evaluating the clinical utility of MRD studies in patients with t (8;21)-containing leukemia. According to the recent follow-up reports using competitive RT-PCR or real-time RT-PCR, the level of the AML1/ETO transcripts in patients in long-term remission became decreasingly convert to negative results and their relapse rate is significantly lower than persistently high-concentration patients25, 26). The quantitative RT-PCR assays for chimeric AML1/ETO transcripts may be very useful for the monitoring of MRD and detecting early relapse, as well as in determining the chemotherapy alone or chemotherapy in conjunction with bone marrow transplantation as intensification treatment of AML.
The AML1/CBF beta transcript factor complex functions as a critical regulator of the development of the adult hemopoietic system. The biological functions controlled by AML1/CBF beta appear to be highly vulnerable to oncogenic mutations, suggesting that alterations in this complex provide an efficient mechanism for driving leukemogenesis. AML1/ETO can function as a dominant inhibitor of normal AML1/CBF beta activity. AML1/ETO expressing cells differentiated abnormally and had a high self-renewal capacity, readily establishing immortal cell lines in culture. AML1/ETO not only represses normal AML1-mediated functions but also generates signals that contribute to the initiation of aberrant hemopoietic cell proliferation27, 28). Future studies aimed at better defining the AML1/CBF beta interactive proteins required for its normal transcriptional activity and identifying the critical downstream targets that are mechanistically involved in its hemopoietic functions should lead to important advances in our understanding of this disease and, hopefully, to the development of effective new therapies. In the future, these methodologies are likely to be incorporated not only into the routine diagnostic evaluation of AML patients, but also into our approaches to monitor a patient's therapeutic response. The information gained from such studies should enhance our ability to individually tailor therapy for each patient and to improve the chances for a cure.
Figure┬Ā1.
Detection of AML1/ETO specific transcripts by RT-PCR in AML patients and cell lines. Lane M: length marker ŽłX174F/Hae III-digest, lane 1 & 4: positive patients, lane 2: K562 cell line, lane 3: primer control (no RNA), lane 5: negative patients.

Figure┬Ā2.
Morphologic finding of AML1/ETO positive and negative patients with AML-M2 (├Ś1,000). (A) AML1/ETO positive patients. There are Auer rod (Ōåæ), abnormal granules in leukemic blast. The size of blasts are large and they have prominent golgi. (B) AML1/ETO negative patients. No blasts have Auer rod, eosinophils, abnormal granules and prominent golgi.
Table┬Ā1.
Primer sequences
| Primer | Sequence | Product | |
|---|---|---|---|
| Outer | sense | 5ŌĆ▓-AGCCATGAAGAACCAGG-3ŌĆ▓ | |
| antisense | 5ŌĆ▓-AGGCTGTAGGAGAATGG-3ŌĆ▓ | 338 bp | |
| Inner | sense | 5ŌĆ▓-CTACCACAGAGCCATCAA-3ŌĆ▓ | |
| antisense | 5ŌĆ▓-TGAACTGGTTCTTGGAGC-3ŌĆ▓ | 185 bp | |
Table┬Ā2.
Comparison of clinical and laboratory findings between AML1/ETO positive and negative group
Table┬Ā3.
Comparison of immunophenotypic findings between AML1/ETO positive and negative group in AML and AML-M2
Table┬Ā4.
Comparison of morphologic characteristics between AML1/ETO positive and negative group within AML-M2
| Positive (n=13) | negative (n=16) | p | |
|---|---|---|---|
| Auer rod | 69.2% | 6.2% | 0.001 |
| Eosinophilia* | 30.8% | 12.5% | 0.364 |
| Abnormal granules | 53.8% | 12.5% | 0.041 |
| Large blast with prominent golgi | 92.3% | 12.5% | 0.001 |
REFERENCES
1. Fenaux P, Preudhomme C, Lai JL, Morel P, Beuscart R, Bauters F. Cytogenetics and their prognostic value in de novo acute myeloid leukaemia: a report on 283 cases. Br J Haematol 73:61ŌĆō671989.


2. Swirsky DM, Li YS, Matthews JG, Flemans RJ, Rees JK, Hayhoe FG. 8;21 translocation in acute granulocytic leukemia: cytological, cytochemicai and clinical features. Br J Haematol 56:199ŌĆō2131984.


3. Nucifora G, Dickstein Jl, Torbenson V, Roulston D, Rowley JD, Vardiman JW. Correlation between cell morphology and expression of the AML1/ETO chimeric transcript in patients with acute myeloid leukemia without the t (8;21). Leukemia 8:1533ŌĆō15381994.

4. Berger R, Bernheim A, Daniel MT, Valensi F, Sigaux F, Flandrin G. Cytologic characterization and significance of normal karyotypes in t (8;21) acute myeloblasts leukemia. Blood 59:171ŌĆō1781982.

5. Kita K, Nakase K, Miwa H, Masuya M, Nishii K, Morita N, Takakura N, Otsuji A, Shirakawa S, Ueda T, Nasu K, Kyo T, Dohy H, Kamada N. Phenotypical characteristics of acute myelocytic leukemia associated with the t (8;21) (q22;q22) chromosomal abnormality: frequent expression of immature B-cell antigen CD19 together with stem cell antigen CD34. Blood 80:470ŌĆō4771992.

6. Paietta E, Wiernik PH, Andersen J. Immunophenotypic features of t(8;21)(q22;q22) acute myeloid leukemia in adults. (Letter:comment) Blood 81:1975. 1993.

7. Bloomfield CD, Lawrence D, Byrd JC, Carroll A, Pettenati MJ, Tantravahi R, Patil SR, Davey FR, Berg DT, Schiffer CA, Arthur DC, Mayer RJ. Frequency of prolonged remission duration after high dose cytarabine intensification in acute myeloid leukemia varies by cytogenetic subtype. Cancer Res 58:4173ŌĆō41791998.

8. Burnett A, Goldstone A, Steven R, Hann L, Rees J, Wheatley K, Gray R. The rote of BMT in addition to intensive chemotherapy in AML in first CR: results of the MRC AML-10 trial. (Abstract) Blood 84:992. 1994.
9. Groupe Francais de Cytogenetique Hematologique (GFHC). Acute myelogenous leukemia with an 8;21 translocation: a report on 148 cases from the Groupe Francais de Cytogenetique Hematologique. Cancer Genet Cytogenet 44:169ŌĆō1791990.
10. Erickson P, Gao J, Chang KS, Look T, Whisenant E, Raimondi S, Lasher R, Trujillo J, Rowley J, Drabkin H. Identification of breakpoints in t (8;21) acute myelogenous leukemia and isolation of a fusion transcript, AML 1/ETO, with similarity to Drosophila segmentation gene, runt. Blood 80:1825ŌĆō18311992.

11. Shimizu K, Miyoshi H, Kozu T, Nagata J, Enomoto K, Maseki N, Kaneko Y, Ohki M. Consistent disruption of the AML1 gene occurs within a single intron in the t (8;21) chromosome translocation. Cancer Res 52:6945ŌĆō69481992.

12. Kim KH, Kim TG, Han JY, Kim JM, Kim JS, Kim HJ, Lee YH, Lee EY. Cytogenetical, morphological, and immunophenotypical characteristics of myeloid malignancies with t (8;21). Korean J Hematology 34:18ŌĆō261999.
13. Lee SB, Kim Yl, Yoon H-J, Kim SY, Cho KS. AML1/ETO fusion gene expression and clinical characteristics of adult acute myelogenous leukemia. Korean J Hematology 33:311ŌĆō3211998.
14. Downing JR, Head DR, Curcio-Brint AM, Hulshof MG, Motroni TA, Raimondi SC, Carroll AJ, Drabkin HA, Willman C, Theil KS, Civin CI, Erickson P. An AML1/ETO fusion transcript is consistently detected by RNA-based polymerase chain reaction in acute myelogenous leukemia containing the t (8;21)(q22;q22) translocation. Blood 81:2860ŌĆō28651993.

15. Langabeer SE, Walker H, Rogers JR, Burnett AK, Wheatley K, Swirsky D, Goldstone AH, Linch DC. Incidence of AML1/ETO fusion transcripts in patients entered into the MRC AML trials MRC Adult Leukemia Working Party. Br J Haematol 99:925ŌĆō9281997.


16. Andrieu V, Radford-Weiss I, Troussard X, Chane C, Valensi F, Guesnu M, Haddad E, Viguier F, Dreyfus F, Varet B, Flandrin G, Macintyre E. Molecular detection of the t (8;21)/AML1-ETO in AML M1/M2: correlation with cytogenetics, morphology and immunophenotype. Br J Haematol 92:855ŌĆō8651996.


17. Mitterbauer M, Kusec R, Schwarzinger I, Haas OA, Lechner K, Jaeger U. Comparison of karyotype analysis and RT-PCR for AML1/ETO in 204 unselected patients with AML. Ann Hematol 76:139ŌĆō1431998.


18. Serriera JE, Albitar M, Estrov Z, Gidel C, Aboul-Nasr R, Manshouri T, Kornblau S, Chang KS, Kantarjian H, Estey E. Comparison of outcome in acute myelogenous leukemia patients with translocation (8;21) found by standard cytogenetic analysis and patients with AML1/ETO fusion transcript found only by PCR testing. Leukemia 15:57ŌĆō612001.


19. Zhang T, Hillion J, Tong JH, Cao Q, Chen SJ, Berger R, Chen Z. AML-1 gene rearrangement and AML1-ETO gene expression as molecular markers of acute myeloblasts leukemia with t (8;21). Leukemia 8:729ŌĆō7341994.

20. Bloomfield CD. Prognostic factors for selecting curative therapy for adult acute myeloid leukemia. Leukemia:6(suppl 4):65ŌĆō671992.

21. Grimwade D, Walker H, Oliver F, Wheatley K, Harrison C, Harrison G, Rees J, Hann I, Stevens R, Burnett A, Goldstone A. The importance of diagnostic cytogenetics on outcome in AML: analysis of 1,612 patients enrolled into the MRC AML 10 trial. The Medical Research Council Adult and Children's Leukaemia Working Parties Blood 92:2322ŌĆō23331998.

22. Saunders MJ, Brereton ML, Adams JA, Tobal K, Liu Yin JA. Expression of AML1/MTG8 transcripts in clonogenic cells grown from bone marrow of patients in remission of acute myeloid leukemia with t (8;21). Br J Haematol 99:921ŌĆō9241997.


23. Miyamoto T, Nagafuji K, Akashi K, Harada M, Kyo T, Akashi T, Takenaka K, Mizuno S, Gondo H, Okamura T, Dohy H, Niho Y. Persistence of multipotent progenitors expressing AML1/ETO transcripts in long-term remission patients with t (8;21) acute myelogenous leukemia. Blood 87:4789ŌĆō47961996.

24. Morschhauser F, Cayuela JM, Martini S, Baruchel A, Rousselot P, Socie G, Berthou P, Jouet JP, Straetmans N, Sigaux F, Fenaux P, Preudhormme C. Evaluation of minimal residual disease using reverse-transcription polymerase chain reaction in t (8;21) acute myeloid leukemia: a multicenter study of 51 patients. J Clin Oncol 18:788ŌĆō7942000.
25. Tobal K, Newton J, Macheta M, Chang J, Morgenstern G, Evans PA, Morgan G, Lucas GS, Liu Yin JA. Molecular quantitation of minimal residual disease in acute myeloid leukemia with t (8;21) can identify patients in durable remission and predict clinical relapse. Blood 95:815ŌĆō8192000.

26. Kondo M, Kudo K, Kimura H, Inaba J, Kato K, Kojima S, Matsuyama T, Horibe K. Real-time quantitative reverse transcription-polymerase chain reaction for the detection of AML1-MTG8 fusion transcripts in t (8;21)-positive acute myelogenous leukemia. Leukemia Res 24:951ŌĆō9562000.

-
METRICS

-
- 10 Crossref
- 0 Scopus
- 13,018 View
- 80 Download
- Related articles
-
Acute Appendicitis in Patients with Acute Leukemia1993 January;8(1)





 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Download Citation
Download Citation Print
Print


