INTRODUCTION
Helicobacter pylori is now recognized as the major pathogenic factor for the development of chronic gastritis type B, peptic ulcer, and is strongly associated with gastric adenocarcinoma and lymphoma. H. pylori infection is a worldwide problem and more than 50% of the world’s population were infected with H. pylori. The prevalence of H. pylori infection in Korea was more than 70% of the population and virulence factors such as cagA or vacA of the H. pylori isolated from Korean adults revealed high levels compared with those from western countries1).
To understand the pathogenesis of H. pylori infection and to develop novel therapies and vaccines, an adequate animal model to reproduce the various aspects of H. pylori disease is required. Early attempts to colonize rodents with H. pylori were unsuccessful2,3). The first models of H. pylori infection were large animals such as gnotobiotic piglets4), monkeys5) and mice which do not express normal immune systems like euthymic germ-free mice6) and athymic nude mice7). These animal models can not be used easily to study immune response or to develop vaccines against H. pylori infection because they are more expense and difficulty for handling than small-sized animals, such as mice. H. felis or H. mustalae, which are different from H. pylori, have been used to infect mice8) or ferrets9), respectively. However, these animal models do not mimic H. pylori infection in man and subsequent pathologic features because those Helicobacter species do not have VacA and other virulence factors required for the induction of inflammation and ulcers10).
Recently, there has been some success in the development of a mouse model using human strains of H. pylori11,12). Unfortunately, there has not been any report about the mouse model infected with H. pylori in Korea. Considering the geographic differences in the prevalence of virulence factors such as cagA or vacA of H. pylori isolated from Korean adults, compared with those from western countries1,13), the establishment of a mouse model infected with H. pylori isolated from Korean adults is needed to investigate the pathogenesis of H. pylori infection and to develop the vaccines. The present study describes the first attempt to establish a mouse model by direct inoculation of fresh H. pylori isolates to specific pathogen-free BALB/c mice without long-term adaptation.
MATERIALS AND METHODS
1. Bacterial strains
H. pylori was isolated from the antral biopsy specimens of patients with duodenal ulcer at Seoul National University Hospital. Biopsy specimens were placed in Brucella broth (Difco Laboratories, Detroit, MI, USA) immediately and homogenized with a tissue grinder. The homogenate was then inoculated on selective agar [GC medium base (Difco Laboratories). 0.024% yeast extract, 1% hemoglobin, 1% IsoVitaleX, 5 mg/L vancomycin, 1 mg/L mycostatin, 5% sheep blood]. The plates were incubated for 5–7 days at 37°C under microaerophilic conditions (5% O2, 10% CO2 and 85% N2) in a CO2 incubator (Napco 5410, Tualatin, Oregon, USA). Pure culture isolates were examined by Gram stain and biochemical assay such as urease test14). Isolates were finally confirmed as members of the genus Helicobacter by a polymerase chain reaction (PCR) as described below. For a long-term storage, isolates were stored in Brucella broth containing 15% (v/v) glycerol and kept at −70°C.
2. Isolation of bacterial DNA
DNA was extracted from the H. pylori isolates with proteinase K, sodium dodecyl sulfate and hexadecyltrimethyl ammonium bromide (Sigma, St. Louis, MO, USA). The cell lysate was extracted in sequential steps with equal volumes of phenol, phenol/chloroform/isoamylalcohol (25:24:1) and chloroform. DNA was then precipitated with isopropanol. The DNA pellet was washed with 70% ethanol, dried and resuspended in sterile Tris-EDTA buffer (pH 8.0).
3. RNA extraction and reverse transcription
Total RNA of H. pylori was extracted by the acid guanidinium thiocyanate-phenol-chloroform method15) (4 M guanidinium thiocyanate, 25 mM sodium citrate, pH 7.0; 0.4% sarcosyl, 0.1 M 2-mercaptoethanol). The RNA was dissolved in diethyl pyrocarbonate-treated distilled water, and quantitated at absorbance of 260 nm. One μg of each RNA of H. pylori isolated from human and mouse was used for single-strand cDNA synthesis with oligo(dT)15 primer (Promega, Madison, WI, USA) and Moloney murine leukemia virus reverse transcriptase (Gibco BRL, Gaithersburg, MD, USA) as described previously16).
4. Polymerase chain reaction (PCR)
A number of PCRs were performed as described previously17) to characterize the H. pylori isolates obtained from humans or mice. These were Helicobacter-specific PCR, ureA, cagA and vacA PCRs and random amplified polymorphic DNA (RAPD) PCR18). Each PCR primer was designed on the basis of published sequences of H. pylori 1,16,19,20) as shown in Table 1. The primer sequence used for RAPD PCR was based on the publication by Lee et al.12) (5′-AACGCGCAAC-3′). Amplification of H. pylori genomic DNA sequences was carried out in a volume of 50 μl containing PCR buffer [50 mM KCl, 10 mM Tris-HCl (pH 8.3)], 1.5 mM MgCl2, 200 M dNTP, 0.5 M primers, 2 U of Taq polymerase (Perkin-Elmer, Norwalk, CT, USA) and 100 ng of bacterial DNA or cDNA. Each reaction mixture was amplified for 33–39 cycles (shown in Table 2). In RAPD PCR, the MgCl2 concentration was increased to 3 mmol/L and 20 pmol of a single primer was used. A sample omitting DNA or cDNA was included in every reaction as a negative control. Each PCR was performed with hot-start procedure21,22) and the final extension was performed at 72°C for 10 min after the completion of the amplification cycles using a thermal cycler (Perkin Elmer). PCR products were separated in 2% NuSieve agarose gel (FMC Bioproducts, Rockland, ME, USA) and identified using ethidium bromide stains.
5. Inoculation of mouse with H. pylori
Twenty-two, specific pathogen-free, 6-week-old female BALB/c mice were divided into two groups (4 of control and 18 of experiment). Three of frozen cagA+/vacA+ H. pylori strains (strain # 99, # 232 and # 234) and one of fresh clinical cagA+/vacA+ H. pylori isolates (strain # 7) were used in this study. Each H. pylori strain was cultured under microaerophilic conditions and was harvested in the sterile phosphate buffered saline (PBS, pH 7.4) and mixed by equal density. Mice were infected with the H. pylori mixture as described previously23). Briefly, all animals were fed on a commercial diet and given water ad libitum. Mice were fasted overnight except for water. Mice were inoculated intragastrically with 109 colony forming units (CFUs) of H. pylori mixture or PBS after treatment with 0.2 ml of 0.2 M NaHCO3 intragastrically to neutralize gastric acidity. These procedures were repeated two or more times with a 2-day interval.
6. Rapid urease test and histopathologic examination of gastric mucosa of mice
Mice were sacrificed 1, 2, 4 or 6 weeks after the last inoculation. The stomach of each mouse was bisected longitudinally. A specimen for rapid urease test (CLO test, Delta West Pty Ltd, Western Australia) was obtained from one half of the stomach and the remains of it were used for gastric scrapings for H. pylori cultures. They were fixed in buffered 10% formalin, using standard procedures, embedded in paraffin, sectioned at 4 ml, and stained with hemotoxylin & eosin for histology and Warthin-Starry silver to assess the level of bacterial colonization24). The glandular mucosae of the body, antrum and pylorus were examined histologically for a variety of inflammatory responses, epithelial changes and the presence of H. pylori.
7. Enzyme linked immunosorbent assay (ELISA)
ELISA was performed for the assessment of immune response in H. pylori-infected mice as described previously25,26) with minor modification. Briefly, to obtain the antigen to coat the microtiter plate, colonies of H. pylori were sonicated for three 30 sec bursts with 30 sec resting periods in an MSE Soniprep 150, and ultracentrifuged at 100,000 × g for 60 min (Beckman TL-100, Palo Alto, CA, USA). The antigen was diluted in 0.1 M carbonate buffer (pH 9.6) to a final concentration of 10 μg/ml. Microtitre plates were coated with 100 μl/well of antigen solution and incubated overnight at 4°C, washed three times with PBS containing 0.05% Tween-20 and then blocked with 200 μl/well of 1% bovine serum albumin in PBS/Tween-20 at room temperature for 2 hours. After washing the plates, 100 μl of each serum sample diluted 1:75 in PBS were added to wells in duplicate and the plates were incubated at room temperature for 2 hours. The plates were then washed three times and incubated with 100 μl/well of a goat anti-mouse IgG peroxidase conjugate (Pierce, Rockford, IL, USA) and diluted 1:700 in 1% BSA-PBS at room temperature for 2 hours. After three washings, 100 μl/well of a substrate solution containing p-nitrophenyl phosphate in diethanolamine-MgCl2 buffer was added to each well. The reaction was stopped with 50 μl/well of 2 N H2SO4, and read the absorbance read using Wellscan microplate reader at OD450 nm (Dynatech MR 700, Alexandria, VA, USA).
RESULTS
1. Infection rate after inoculation
Specific pathogen-free BALB/c female mice were inoculated with H. pylori. Mice were sacrificed 1, 2, 4 and 6 weeks after the last inoculation. Gastric specimens from infected mice were processed for urease test and histologic evaluation. The positivity of CLO tests in gastric specimens obtained from mice sacrificed one and two weeks after the last inoculation was only 40% but 75% in those from 4 and 6 weeks. (Table 3). As presented the above, the positivity of CLO test was gradually increased from 4 weeks after treatment with H. pylori, which indicates that the colonized H. pylori proliferated slowly and was located in the stomach. The CLO test in the control group was negative during the experimental period.
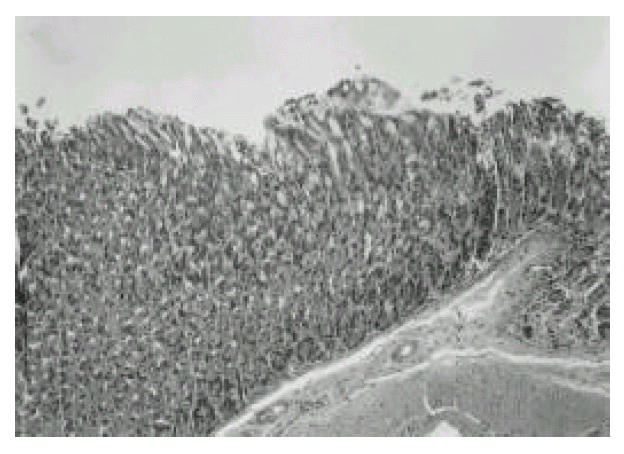
2. Histologic evaluation of gastric mucosa of mice infected with H. pylori
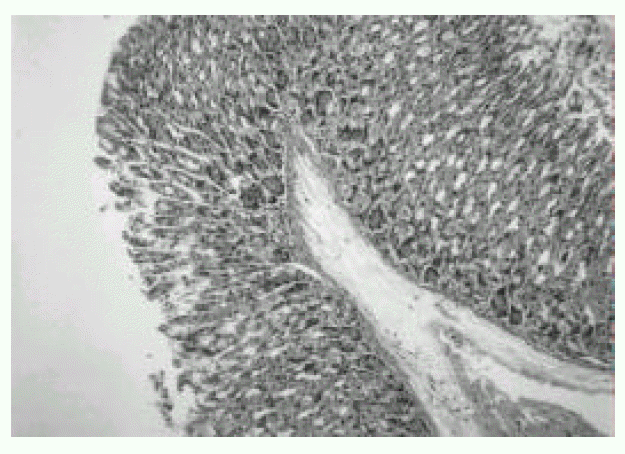
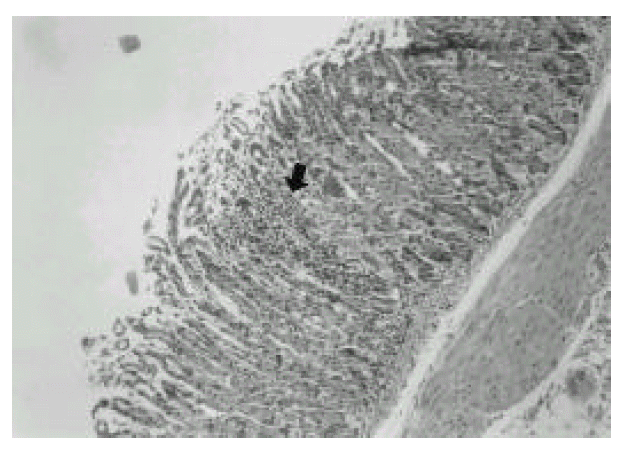
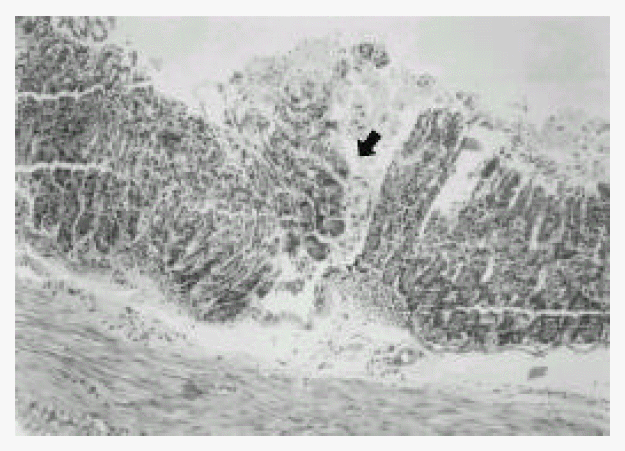
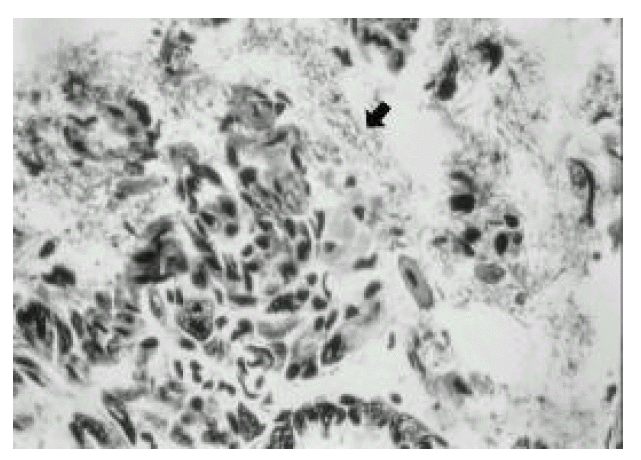
In the first week after the last inoculation, no definite presence of bacteria and infiltration of inflammatory cells were shown in H. pylori-infected group (Fig. 1). However, the inflammatory infiltrates of lymphocytes, plasma cells and polymorphonuclear leukocytes in the antrum and corpus mucosa were gradually increased from 2 weeks after the last inoculation of H. pylori. At six weeks after the last inoculation, the gastric lesions were characterized by definite inflammatory infiltrates accompanied by the disruption of gastric architecture (Fig. 2, 3) and distinct H. pylori colonization (Fig. 4). Control mice showed no evidence of significant infiltration of inflammatory cells during the entire experimental period (Fig. 5).
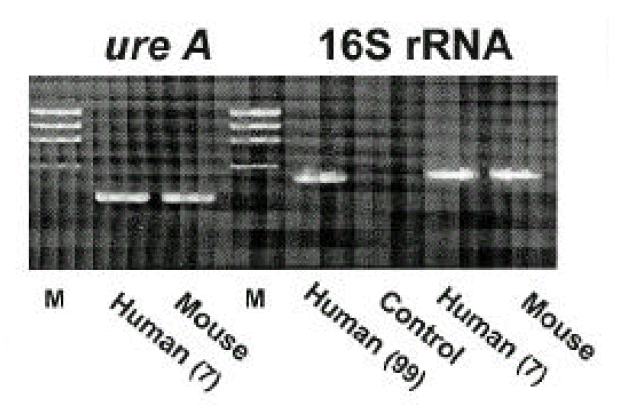
3. Bacteria culture and PCR
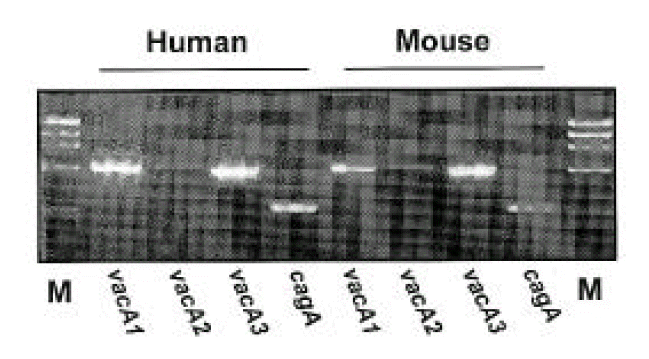
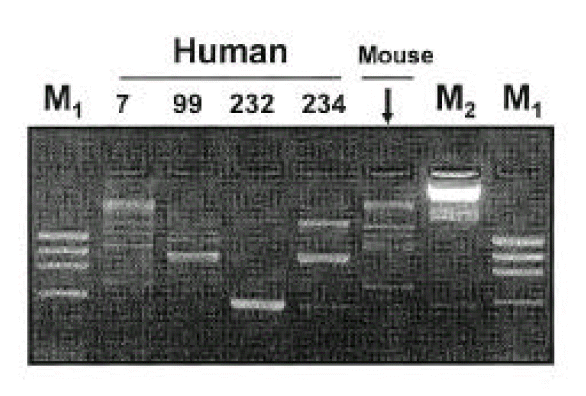
Isolates from infected mice showed typical morphology and biochemical characteristics of H. pylori as the same as the original H. pylori strain (data not shown). Furthermore, the Helicobacter-specific PCR showed that the mouse and human isolates were identical to H. pylori (Fig. 6). The original human clinical isolates and the mouse isolates were also positive for cagA and vacA by PCR (Fig. 7). A comparison of the genomic DNA by RAPD PCR was performed for the mouse isolates and four original human clinical isolates which were used to infect the mouse. The mouse isolates gave an identical band pattern to only one (strain # 7) of the four human isolates (Fig. 8). These results suggest that strain # 7 may be the one that is able to colonize the mouse gastric mucosa among all the human isolates tested.
4. Serum antibody response to H. pylori infection
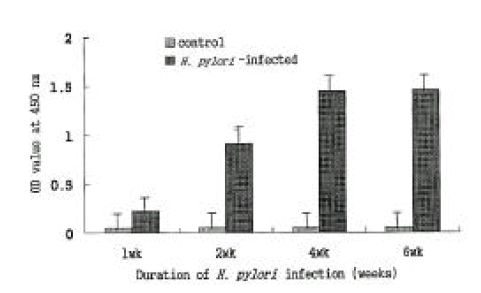
The experimentally infected mice showed serum antibody response to the colonizing strain that could be detected by an ELISA of mice sera from the first week of infection. Anti-H. pylori IgG antibody levels increased markedly from the second week after the last inoculation with H. pylori and reached the plateau after the fourth week. In contrast, the negligible serum antibody response was shown in the control group during the entire experiment period (Fig. 9).
DISCUSSION
H. pylori has been unambiguously implicated in the etiology of chronic gastritis and the recurrence of peptic ulceration in humans. Despite the high prevalence of H. pylori infection, serious gastric diseases characterized by gastric or duodenal ulcers are noted in a relatively small fraction of the H. pylori-infected population: To understand how H. pylori infects and occasionally causes serious diseases, and to investigate the immunologic mechanisms of H. pylori infection more precisely, it is necessary to develop the animal model infected with H. pylori. In the present study, we successfully established an animal model by oral challenges with a fresh H. pylori isolate to BALB/c mice without a long-term adaptation. Based on the result of urease tests, more than a half of the challenged mice were presumed to be infected by H. pylori. A striking feature of infection was the increasing serum IgG response against H. pylori sonicate antigen; infected mice showed a systemic antibody response to the infected strain from the first week of infection. Serum IgG response was apparent in all mice by 6 weeks post-infection although the CLO positive was only 55% in all infected mice. We speculate that it may be due to a peculiar colonization pattern of H. pylori in gastric mucosa. H. pylori has acquired particular properties of colonization of the unique ecological niche on the surface of gastric epithelial cells and the distribution of H. pylori with associated inflammation are often patchy27,28). Such a patchy distribution of H. pylori colonization can lead to sampling error and subsequently, the false negative results of microscopic examination, culture and rapid urease test. Furthermore, the process of colonization with H. pylori in the gastric mucosa may take week29). The organism must enter the stomach, survive brief exposure to acid, traverse the mucous layer30), attach to epithelial cell receptors31), adapt its physiology to the hostile host environment and thereby establish its niche.
In this study, only the bacteria of a negligible number were found in the gastric antrum. However, the colonization with numerous bacteria was observed in the body and fore-stomach transition zones, accompanied with the disruption of gastric gland architecture. With light microscopy, the bacteria were observed in large numbers in the mucus overlying the epithelial cells and at the top of the gastric pits. It is closely similar to the colonizing pattern of H. pylori in humans. The infiltration of inflammatory cells, such as lymphocytes and plasma cells, in the antrum and corpus mucosa was gradually increased from 2 weeks after the last inoculation of H. pylori. In contrast, the infiltration of polymorphonuclear leukocytes was not prominent in the lamina propria until 4 weeks post-infection. However, it became evident at 6 weeks, although with only a few numbers.
In the present study, both H. pylori isolates from human (before inoculation to mouse) and mouse showed identical patterns of virulence factors such as vacA1, vacA2, vacA3 and cagA. By RAPD PCR, the mouse isolate exhibited an identical band pattern to only one (fresh isolate # 7) of the four human isolates. This result suggests that the fresh isolate plays an important role in the colonizing process compared to long-term stored strains. Consecutive inoculation of ICR mice with H. pylori recovered from the stomach of BALB/c mice also showed systemic antibody response to the colonizing strain (data not shown).
The present study shows that a mouse model of H. pylori infection is successfully established. This model can be utilized for animal experiments of H. pylori, such as vaccine studies, screening for novel therapies, and investigation of the mechanisms of pathogenesis. Although some problems remain to be solved, such as to develop the strain with high colonizing ability and to examine a variety of mice strain, this mouse model will provide opportunities for studies on the interrelationship between bacteria and the host with respect to colonization and the ecology of bacteria in the stomach. It will also facilitate investigations of the mechanisms of H. pylori-associated diseases, including peptic ulcer, gastric cancer and gastric lymphoma.



 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Download Citation
Download Citation Print
Print





