 |
 |
| Korean J Intern Med > Volume 3(1); 1988 > Article |
|
Abstract
A dot blot hybridization technique utilizing a biotin-labelled recombinant DNA probe was used to examine hepatitis B virus (HBV) DNA in serum. The lowest amount of HBV DNA in serum detectable by the color development of an avidin-biotin alkaline phosphatase complex was 40 picogram per 50 microliter. Validity of this method was confirmed by autoradiography using 32P-labelled and 3H-labelled HBV DNA probes. HBV DNA was found in 100% (34/34) of the HBsAg-positive and in 73.5% (25/34) of the HBsAg-negative subjects. In contrast, all nine cases showing negativity in HBV DNA were also HBsAg-negative. Correlation of HBe antigen/antibody with HBV DNA was investigated in 19 sera of which HBsAg was negative but anti-HBc-positive. Of 13 sera with anti-HBe 10 (76.9%) cases revealed HBV DNA positivity, while four (66.7%) of six sera without anti-HBe were positive in HBV DNA.
In conclusion, serum dot hybridization assay utilizing a biotinylated probe proved useful in the detection of a free form of HBV DNA regardless of the presence of HBsAg and irrespective of HBeAg/anti-HBe status. Moreover, it is emphasized that practical advantages in speed, reproducibility, and safety have made this alternative even more attractive than autoradiography using radioisotope-labelled probes.
The status of HBV infection has been serologically determined by radioimmunoassay for HBV-associated antigens and their antibodies. Persistant replication of HBV, however, can be demonstrated in liver tissue sections even after complete disappearance of HBsAg in serum where seroconversion of HBeAg to anti-HBe has occurred.1) The recent availability of recombinant DNA technology and molecular hybridization techniques now permits detection of HBV DNA in serum to the picogram level based on DNA sequence homology.2ŌĆō7) The quantitative determination of HBV DNA in the serum by dot blot hybridization methods has been shown to be both specific and sensitive, indicating the level of replication of HBV DNA. The most widely used methods of dot blot or moleculer hybridization employ autoradiography to detect HBV DNA as radioactive signals. The disadvantages of using these techniques include the short half-life of the radio-labelled probes, radiation hazard, and the need for exposure time to produce detectable signals in the x-ray film.
The use of immunological and enzymatic methods for the detection and quantitation of specific macromolecules has become increasingly widespread in molecular and cell biology. In particular, many experimental approaches call for the ability to reliably detect nanogram amount of proteins immobilized on nitrocellulose paper, either by western blot or dot blot.
In this study, we intended to detect HBV DNA in serum by dot blot hybridization using a biotinylated nucleotide probe in which the signal was demonstrated by an enzymatic reaction of an avidin-biotin-alkaline phosphatase complex linked to the target DNA. The correlation of HBV- associated antigen-antibody markers with HBV DNA in serum was established and analyzed.
Serum samples were obtained from 59 subjects with various liver diseases and/or serological evidence of present or past HBV infection. Sera was taken also from six candidates presenting negative in all HBV-associated serological markers as the negative control. These 65 sera were stored at ŌłÆ20┬░C until being tested blindly.
Escherichia coli strain HB 101 containing recombinant pHBV 315 DNA was kindly provided by Dr.M.H.Yu (Genetic Engineering Center, KAIST). This plasmid contains the full-length HBV DNA as well as pBR 322 sequences. The bacterial cells were grown in 100 ml LB broth containing ampicillin (Bactotrypton 1%, Bacto-yeast extract 0.5%, NaCl 1%, pH 7.5, ampicillin 0.0035%) overnight with vigorous shaking. The plasmid was isolated by the modified method of Birnboim and Doly.8) The cells were harvested at 4┬░C by spinning at 7,000 ├Ś g for 10 minutes. Ten ml of iceŌĆöcold solution I (50 mM glucose, 25mM Tris-HCl, pH8.0, 10mM EDTA) was added to resuspend the cell. The cells were harvested again, and resuspended with 3 ml of ice-cold solution I containing 4mg/ml lysozyme. Following incubation for 10 minutes at room temperature, solution II (0.2N NaOH, 1% SDS) was added and gently mixed. After 10 minutes on ice, 4.5 ml of cold 5M potassium acetate (pH 4.8) was added, and white precipitates developed. These were centrifuged at 9,000├Śg at 4┬░C for 20 minutes. The supernatant was collected and extracted with phenol. The aqueous phase was separated and precipitated with ethanol.
Instead of using ultracentrifugation for purification of plasmid DNA we employed the PEG precipitation method,9) which is simpler and faster.
Purified pHBV 315 DNA was biotinylated by the nick translation method of Rigby et al.10) Two ╬╝g of probe DNA was mixed in 50 ╬╝l of solution containing 50 mM Tris-HCl, pH7. 8, 10mM MgCl2, 50╬╝g/ml bovine serum albumin, 0.1 mM DTT, 20 ╬╝M of dATP, dGTP, dCTP (Sigma, St. Louis, MO.) and bio-11-dUTP (Bethesda Research Lab, Gaithersburg, MD.). In order to create nicks in the DNA, this mixture was treated with 0.2 ng of pancreatic DNase I (Sigma, St. Louis, MO.) at 37┬░C for 15 minutes. Four to five units of E. Coli DNA polymerase I (Pro-mega, Madison, WI.) were added and the reaction proceeded at 15┬░C for 3.5 hours. The reaction was stopped by adding 2╬╝l of 0.5 M EDTA.
In a parallel reaction, 32P-labelled HBV DNA probe and 3H-labelled probe were prepared by adding either alpha-32P dATP (3,000 Ci/mM) (New England Nuclear) or 3H dATP (100 Ci/mM) instead of bio-11-dUTP in a solution containing 20 ╬╝M of the other 3 NTPŌĆÖs.
Labelled HBV DNA was purified by precipitating twice with ethanol in sodium salt and ammonium salt.
Fifty ╬╝l of cleared serum samples and 50 ╬╝l of unlabelled probe of serially diluted concentration markers were treated with 1 M NaOH and 2M NaCl for 10 minutes, and then filtered through nitrocellulose paper mounted on the Hybridot system (BRL Inc. Gaithersburg, MD.) under vacuum. For neutralization, 200 ╬╝l of solution containing Tris-HCl, pH 7.4, 3M NaCl, were filtered again. The air-dried nitrocellulose filter paper was baked at 80┬░C in a vacuum oven for 2 hours. Prehybridization of the filter paper was carried out at 42┬░C for 1.5 hours in 50% deionized formamide 5├Ś SSC (1├ŚSSC = 0.015M sodium citrate, 0.15M NaCl), and DenhardtŌĆÖs medium (0.1% ficoll, 0.1% BSA, 0.1% polyvinyl pyrrolidone) in a polyethylene bag. The filter was hybridized at 42┬░C for 18 hours with 3ml of 2├ŚSSC containing labelled HBV DNA probe at a concentration of 0.5ng/ml. It was then washed three times in 0.2├ŚSSC at room temperature and once at 40┬░C.
For detection of the biotinylated HBV DNA on the filter, the nitrocellulose was incubated for 15 minutes at 42┬░C in TTBS (0.1 M Tris-HCl, pH7.5, 0.1 M NaCl, 2mM MgCl2, 0.05% Triton X-100) containing 3% crystalline grade BSA (Sigma, St. Louis, MO.). The air-dried nitrocellulose was baked at 80┬░C for 30 minutes, then rehydrated in TTBS containing 3% crystalline BSA for 20 minutes. The nitrocellulose paper was incubated in avidine-biotin-complex-alkaline phosphatase (Vector Lab. Burlingame, CA.) for 30 minutes in the dark. Color development was carried out by incubation of nitrocellulose in a freshly prepared mixture of substrates of BCIP (5-bromo, 4-chloro, 3-indolyl phosphate) and NBT (nitro blue tetrazolium). Following color development which usually takes 20 minutes to I hour, nitrocellulose was washed with 3 changes TTBS over 10 minutes and then baked at 80┬░C in a vacuum oven.
Signal detection of radioisotope-labelled HBV DNA was performed by autoradiography of the filter using Kodak X-OMAT cassetts with intensifying screen and Kodak XAR-5 film at ŌłÆ80┬░C. Exposure time for developing the film was 24 to 72 hours for the 32P-labelled HBV DNA and 4 weeks for the 3H-labelled probe, respectively. Signal detection of the 3H-labelled HBV DNA was enhanced by using autoradiography enhancer (New England Nuclear).
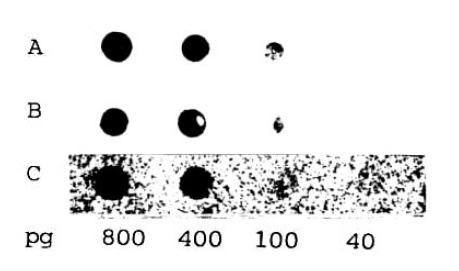
In order to determine the sensitivity of the biotin-labelled HBV DNA probe assay, a control study using a radioisotope-labelled DNA probe was performed. The indicated amounts of pure unlabelled HBV DNA with serial dilutions were spotted directly on nitrocellulose paper in 50 ╬╝l volumes, followed by hybridization with labelled DNA and the signal detection by either enzymatic color development of ABC-AP with substrate or autoradiography of 32P or 3H-labelling. As seen in Figure 1, the 32P-labelled probe is the most powerful in resolution and quantitation of the lowest value of 40pg/50 ╬╝l The tritiated hybridization assay, however, yields poor in resolution to quantitate that value. By dotting recombinant lambda-HBV DNA to nitrocellulose and hybridizing with our biotinylated probe, a standard pattern was established and used to classify the quantity in serum according to the intensity of staining (Figures 1 and 2). The lowest detection limit of the biotinylated probe hybridization assay was found to be also 40pg/50╬╝l but faint in intensity in comparison to the 32P-labelled DNA hybridization assay. Contrarilly, the detection limit of tritiated probe was 100pg/50╬╝l.
Of 31 HBsAg positive sera HBV DNA was positive in all of the samples (100%). Twenty five (73.5%) of 34 HBsAg negative sera were also positive in HBV DNA by dot blot hybridization utilizing the biotin-labelled probe (Table 1). Contrarily, nine sera revealed negative HBV DNA were also negative in HBsAg (Table 2).
Surprisingly, as seen in Table 3, all four sera with positive anti-HBs showed positive results in the HBV DNA assay. In fact, two of these four cases were sera from individuals whom had previously been vaccinated against HBV. Furthermore, half of the six sera which were negative in HBV-associated serological markers displayed positivity of HBV DNA.
Correlation of HBeAg/anti-HBe with HBV DNA was investigated in 19 sera of which HBsAg was negative but anti-HBc-positive. Of 13 sera with anti-HBe 10 (76.9%) cases revealed HBV DNA positivity, while four (66.7%) of six sera negative in anti-HBe were positive in HBV DNA (Table 4). This indicated, in turn, that HBV DNA could be detected regardless of the presence of HBsAg and irrespective of HBeAg/anti-HBe status.
The recent introduction of nonradiographic methods of probe detection has been applied successuflly to investigate molecular events using the Soutern, northern, western or dot blot hybridization protocols.11ŌĆō13) Much of the success of the enzymatic methods can be attributed to the use of substrates that are converted to intensely colored compounds which are precipitated by the reaction with enzyme activity. Instead of the potentially carcinogenic diaminobenzidine tetrahydrochloride and hydrogen peroxide for horse-radish peroxidase (HRP), the combination of BCIP and NBT is just as sensitive substrate for alkaline phosphatase (AP). This utility was applied in the signal detection of biotin-lebelled polynucleotides as well as of immunoblotting. In this reaction, the enzyme converts BCIP to the corresponding indoxyl compound, which precipitates and tautomerizes to ketone. Under the conditions of the assay, the ketone undergoes oxidation and dimerizes to form an indigo, which is blue. The hydrogen ions released from dimerization in turn reduce the NBT salt to the corresponding diformazan, which is intensely purple. This compound is deposited along with the indigo by incubation with avidin-biotinylated AP activity, thus amplifying the signal obtained from a given amount of enzyme without affecting background levels.
Although AAAF (N-acetyl-2-acetylaminofluorene)-labelled HBV DNA was recently reported13) to be a useful probe detected by anti-AAAF monoclonal antibody and incubation with AP-labelled bridging IgG antibody and substrate complex of BCIP and NBT, AAAF is a potential carcinogen. Since the biotin-labelled DNA probe is not difficult to prepare and is stable for months, this is a suitable means for routine testing and despite less sensitivity it provides a major alternative to conventional HBV DNA detection using a 32P-labelled probe of a short functional half life. In addition, it avoids the potential hazard of irradiation. Moreover, it is possible to screen up to 100 samples within 24 hours.
Serum HBe Ag has been thought to be a useful marker reflecting infectivity and active replication of HBV. In our study, however, there were discordant results between serum HBV DNA and the HBeAg/anti-HBe status. Surprisingly, anti-HBs-positive sera exclusive of the other serological markers also yielded HBV DNA even in the two individuals immunized previously with vaccin against HBV. In this respect, our finding is consistent with other reports.14ŌĆō20) This dissociation between the HBV-associated serological markers and HBV DNA assay is supposed to reflect variations in HBV expression with considerably different possibilities.20) For many anti-HBc and anti-HBs-positive patients, the HBsAg might be hidden in immune complexes, the viral particles being only identified by hybridization assay. Alternatively, restricted HBV gene expression in an infected hepatocyte can be due to either lower transcription or modification in the assembly and/or exportation of viral particles. As in our study, it seems now apparent that HBV DNA can be detected in HBsAg-negative sera. However, the prevalence of HBsAg-negative HBV DNA-positive cases may be different in global geographic areas.
In summary, serum dot hybridization assay utilizing biotin-labelled HBV DNA proved useful to detect HBV DNA regardless of HBsAg and irrespective of HBeAg/anti-HBe status. Also an anti-HBs does not always correspond to the clearance of HBV. This indicates that the potential infectivity of certain sera previously considered to be safe should be kept in mind.
Notes
This work is supported by a fund donated by an individual who wishes to remain anonymous. We hereby appreciate his contribution to medical science research.
Fig.┬Ā1.
Comparison of sensitivities of the dot blot hybridization techniques for the quantitation of HBV DNA. The indicated amounts of pure unlabelled HBV DNA were spotted directly on nitrocellulose filter paper in 50 ╬╝l volumes, followed by hybridized either with biotinylated HBV DNA probe (A) or with a 32P-labelled DNA (B) or a 3H-DNA probe (C). After enzymatic reaction (A) or autoradiography (B and C), the detection limit of dot blot assay was found to be 40 pg/50 ╬╝l.
Table┬Ā1.
HBV DNA detection rate by dot blot hybridization of serum using a biotinylated probe
| Radioimmunoassay of HBsAg | HBV DNA Positivity | |
|---|---|---|
| HBsAg Positive | 31 Cases | 31/31 (100.0%) |
| HBsAg Negative | 34 Cases | 25/34 (73.5%) |
|
|
||
| Total | 65 Cases | 56/65 (86.2%) |
Table┬Ā2.
HBV serological markers in 9 cases showing HBV DNA negativity
Table┬Ā3.
Profile of HBV serological makers and HBV DNA positive rate
REFERENCES
1. Hino O, Kitakawa T, Koike K, Kobayahi M, Hara M, Mori W, Nakashima T, Hattori N, Sugano H. Detection of hepatitis B virus DNA in hepatocellular carcinomas in Japan. Hepatology 4:90. 1984.


2. Kam W, Rail LB, Smuckler EA, Schmid R, Rutter WJ. Hepatitis B viral DNA in liver and serum of asymptomatic carriers. Proc Natl Acad Sci USA 79:7522. 1982.



3. Imazeki F, Omata M, Yokosuka O, Matsuyama Y, Ito Y, Okuda K. Analysis of DNA polymerase reaction products for detecting hepatitis B virus in serum-comparison with spot hybridization technique. Hepatology 5:783. 1985.


4. Brechot C, Hadchovel M, Scotto J, Degos F, Charnay P, Trepo C, Tiollais P. Hepatitis B virus DNA in the liver and serum: a direct appraisal of the chronic carrier state. Lancet 2:765. 1981.


5. Berninger M, Hammer M, Hoyer B. An assay for the detection of the DNA genom of hepatitis B virus in serum. J Med Virol 9:57. 1982.


6. Lieberman HM, LaBrecque DR, Kew MC, Hadziyannis SJ, Shafritz DA. Detection of hepatitis B virus DNA directly in human serum by a simplified molecular hybridization test: comparison to HBeAg/anti-HBe status in HBsAg carriers. Hepatology 3:285. 1983.


7. Weller IVD, Fowler MJF, Mongardino J. The detection of HBV-DNA in serum by molecular hybridization: a more sensitive method for the detection of complete HBV particles. J Med Virol 9:273. 1982.


8. Birnboim HC, Doly J. A rapid alkaline extraction procedure for screening recombinant DNA. Nucleic Acids Res 1:1513. 1979.



9. Lee YH. An efficient purification of plasmid DNA from E coli. Korean Biochemistry News Letter 7:10. 1987.
10. Rigby WJ, Dieckmann M, Rhodes C, Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol 133:237. 1977.

11. Leary JJ, Brigati DJ, Ward DC. Rapid and sensitive colorimetric method for visualizing biotin-labeled DNA probes hybridized to DNA or RNA immobilized on nitrocellulose: bio-blots. Proc Natl Acad Sci USA 80:4045. 1983.



12. Langer PR, Waldrop AA, Ward DC. Enzymatic synthesis of biotin-labeled polynucleotides: novel nucleic acid affinity probes. Proc Natl Acad Sci USA 78:6633. 1981.



13. Chollet A, Kawashima EH. Biotin-labeled synthetic oligodeoxyribo-nucleotides: chemical synthesis and uses as hybridization probes. Nucl Acids Res 13:1529. 1985.



14. Larzul D, Thiers V, Courouce AM, Brechot C, Guesdon JL. Non-radioactive hepatitis B virus DNA probe for detection of HBV-DNA in serum. J Hepatology 5:199. 1987.

15. Lok ASF, Hadziyannis SJ, Weller IVD, Karvountzis MG, Monjardino J, Karayiannis P, Montaus L, Thomas HC. Contribution of low level HBV replication in continuing inflammatory activity in patients with anti-HBe positive chronic hepatitis B virus infection. Gut 25:1283. 1984.



16. Bonino F, Rosina F, Rizzetto M, Rizzi R, Chiaberge E, Tardanico R, Callea F, Verme G. Chronic hepatitis in HBsAg carriers with serum HBV-DNA and anti-HBe. Gastroenterology 90:1268. 1986.


17. Feinman SV, Guha A, Berris B. The presence of HBV DNA in asymptomatic anti-HBe positive carriers. Hepatology 4:1052. 1984.
18. Scotto J, Hadchovel M, Hery C, Yvart J, Tiollais P, Brechot C. Detection of hepatitis B virus DNA in serum by a simple spot hybridization technique: comparison with results for other viral markers. Hepatology 3:279. 1983.





 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Download Citation
Download Citation Print
Print



