 |
 |
| Korean J Intern Med > Volume 18(2); 2003 > Article |
|
Abstract
Background :
Although there are many methods including AFB smear and culture, and the analysis of pleural fluid in the etiological diagnosis of pleural effusion, it is sometimes difficult to confirm a diagnosis especially in cases of incomplete pleural biopsies. Moreover, the high incidence of tuberculous pleuritis in young people caused confusion in the differential diagnosis of pleural effusion in Korea. The pathognomonic finding of tuberculous pleuritis in pleural biopsy is chronic granulomatous pleuritis (CGP) with caseous necrosis. But a biopsy does not always provide a definitive diagnosis, which shows in only 60ŌĆō70% of all biopsies, because of either limitations in blind biopsies or inadequate specimens. An adequate biopsy also gives only limited information, such as chronic or nonspecific pleuritis.
Methods :
We compared the clinical diagnosis, pathologic findings and detection of mycobacterial DNA using nested PCR of pleural biopsy tissues. We carried out the nested PCR for IS6110 insertion sequence of Mycobacterium tuberculosis using outer primer IS-1/IS-2 (5ŌĆ▓-AGGCGTTGGTTCGCGAGGG-3ŌĆ▓ / 5ŌĆ▓-TGATGACGCCCTCGTTGCC-3ŌĆ▓) and inner primer IS-3/IS-4 (5-CCAACCCGCTCGGTCTCAA-3ŌĆ▓ / 5ŌĆ▓-ACCGATGGACTGGTCACCC-3ŌĆ▓) in 52 pleural biopsy tissues which were pathologically diagnosed as tuberculous pleuritis, malignant pleuritis or non-specific pleuritis.
Results :
Five (71.4%) of 7 cases clinically and pathologically confirmed tuberculous pleuritis diagnosed as chronic granulomatous pleuritis (CGP) with caseous necrosis revealed positive in nested PCR for M. tuberculosis. Seven (36.8%) of 19 cases diagnosed as CGP without caseous necrosis were positive. However, only 3 (25%) of 12 cases diagnosed as non-specific chronic pleuritis were positive by PCR for M. tuberculosis. Neither congestive heart failure nor malignancies with pleurisy showed a positive reaction.
Conclusion :
In this study, pathologic findings were significantly associated with the detection rate of mycobacterial DNA. And, even in patients with nonspecific or chronic inflammatory pleuritis, mycobacterial DNA could be detected by using nested PCR in pleural biopsy tissue with good specificity. Detection of mycobacterial DNA in pleural tissue might provide additional information for etiological diagnosis in patients with pleural effusion.
Pleuritis is one of the most common forms of extrapulmonary tuberculosis. Tuberculous pleuritis can be diagnosed by clinical findings, rhoentgenography of chest, Mantoux test and analysis of pleural fluid, and confirmed only by pleural biopsy or mycobacterial stain and culture of pleural fluid1). However, a pleural biopsy does not always provide a definitive diagnosis because of a difficult biopsy technique, inadequate specimens and/or limited information, such as chronic or nonspecific pleuritis2ŌĆō4).
Recently, polymerase chain reaction (PCR) of mycobacterial DNA in pleural effusion has been used with variable sensitivity, 25ŌĆō80% in the diagnosis of tuberculous pleural effusion5ŌĆō8). We carried out the nested PCR in pleural biopsy tissue to compare the clinical diagnosis, the pathologic findings and the detection of mycobacterial DNA using nested PCR.
The purposes of this study are first, understanding the diagnosis and differential diagnosis of tuberculous pleuritis from other causes of pleural effusion, especially in cases of an inadequate pathologic diagnosis, such as chronic non-specific pleuritis; second, comparing the pathologic findings to the PCR results and other laboratory findings of pleural fluid; third, studying the correlation between the pathologic findings and the PCR results with or without radiological parenchymal infiltrations.
Tissues were obtained from 52 patients who underwent diagnostic pleural biopsies with Cope needle from Jan 1994 through Dec 1995 at Seoul Paik Hospital, Inje University. Five slices of 7 ╬╝m-thick paraffin-embedded tissue specimens were taken from each pleural biopsy specimen. The blade of the microtome and the toothpick were changed every time to avoid cross-contamination of samples. The sections for PCR were collected in 1.5 mL Eppendorf tubes. Deparaffinization was done with xylene and octane 3 times. The study subjects were divided into 3 groups: subjects with chronic granulomatous pleuritis (CGP) with caseous necrosis (group A), CGD without caseous necrosis (group B) and subjects with chronic or nonspecific pleuritis (group C).
Tissue was obtained from three patients who had congestive heart failure (group D) and eleven patients who had pleural effusion due to malignant tumors (group E), such as lung cancer, mesothelioma, mesenchymal tumor and metastatic lymphoma.
The modified technique for isolation of mycobacterial DNA described by Patel et. al.9) was done. Briefly, the deparaffinized pellet was resuspended in a 200 ╬╝L digestion buffer, 500 ╬╝L of 1 M TRIS-HCI (pH=8) containing 100 mM EDTA, 150 mM NaCI, and 200 ╬╝g/mL of protease K (Sigma Chemical Co., St. Louis, MO) and then we disaggregated the tissue with a 23 gauge syringe.
Next, we incubated it for 1 hour at 55┬░C and overnight at 37┬░C. Then we spun the tubes briefly and removed any liquid. We incubated it at 95┬░C for 10 min to inactivate protease. Then the nucleic acid was purified by phenol/ chloroform extraction and precipitated with isopropanol. The precipitate was redissolved, digested with 4 U/mL RNase. DNA was re-extracted with phenol/chloroform and precipitated with isopropanol repeatedly 3 times. DNA precipitates were dissolved in 10 mM TRIS-HCI (pH=8).
Oligonucleotides were synthesized using a DNA synthesizer (Perceptive) and purified by ethanol precipitation. The sequences for oligonucleotides used in this study are summarized in Table 1. The primers were selected by using the oligomer program to amplify a fragment of the IS6110 insertion element present in the M. tuberculosis complex. DNA amplification was performed as previously described. All reaction contained 10 mM TRIS-HCI (pH 8.3, Sigma), 50 mM KCI, 100 ╬╝g/mL gelatin, 25 pmol each oligonuclotide primer, 0.3 mM each dNTP (Pharmacia), 25 to 50 U/mL Taq polymerase (Bioneer).
The presence of amplified mycobacterial DNA sequences was detected as previously described10ŌĆō15). Briefly, aliquots of the amplification products were electrophoresed into 2% agarose gels. The gels were stained with ethidium bromide and illuminated with ultraviolet light and the presence of visible amplification products was detected by Polaroid camera. The positive control for detection of Mycobacterial DNA was H37Rv strain of M. tuberculosis, and negative control was Mycobacterium avium.
Chest PA and both lateral decubitus radiographs were obtained from all individuals. Two radiologists and one pulmonologist read the radiographs in a random manner by the diagnostic guideline of pulmonary tuberculosis according to the Korean National Tuberculous Association criteria without clinical information. Pulmonary tuberculosis was considered when 2 or more investigators interpreted apical parenchymal infiltrations without a mention of activity.
Blood and pleural fluid were obtained from all individuals at the same time as the pleural biopsy. Adenosine deaminase (ADA) was determined enzymetically using an analyzer (U/L, Hitach 7150); CA125 was measured directly by radioimmunoassay (U/mL, Cobra RIA/QC 5005, 5010). Total protein of serum and pleural fluid were measured by Biurep method (g/dL, Olympus A5400).
The correlation between the pathologic findings, the pleural fluid analysis, the PCR result of mycobacterial DNA and the various clinicoradiologic findings were analysed for statistical significance by the t-test, 2 by 2 Žć 2-test. The statistical software program used was Statistical Package for the Social Sciences (SPSS/PC+ 10.0, Chicago, IL). A p-value of less than 0.05 was accepted as statistically significant.
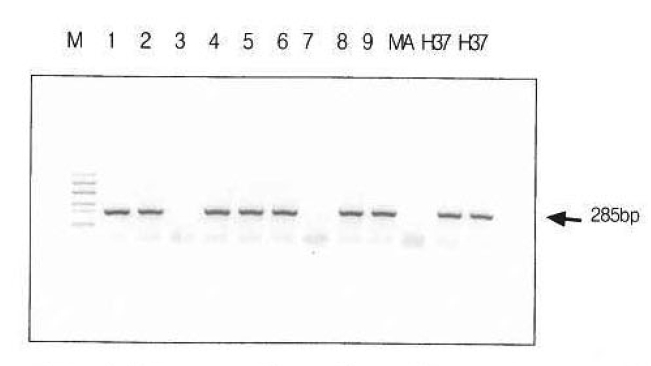
Fifteen (39.5%) of 38 specimens revealed positive in nested PCR for IS6110 insertion sequence of M. tuberculosis. Positive control was the H37Rv strain of M. tuberculosis (H37) and negative control was M. avium (MA) (Figure 1). Five (71.4%) of 7 cases, clinically and pathologically confirmed tuberculous pleuritis diagnosed as chronic granulomatous pleuritis (CGP) with caseous necrosis, revealed positive in nested PCR for IS6110 insertion sequence of M. tuberculosis. Seven (36.8%) of 19 cases diagnosed as CGP without caseous necrosis were positive. However, three (25%) of 12 cases diagnosed as chronic or nonspecific pleuritis were positive in PCR for M. tuberculosis. None of congestive heart failure and malignancies-induced pleurisy showed a positive reaction (Table 2).
When the parenchymal infiltrations in simple chest X-ray suspected of active or inactive pulmonary tuberculosis were combined, positive PCR reaction was higher than without radiological parechymal infiltraitons (p<0.05) (Table 3). This finding suggests that the pathogenesis of tuberculous pleurisy is more likely the rupture of a subpleural focus of tubercle bacilli into the pleural space than an immunologic response to mycobacterial antigen exposure. However, a negative PCR result was not correlated with negative radiological infiltrations. Because of the small number of patients in each group, we could not find any correlation between positive or negative PCR reaction and radiological infiltrations in each group.
Ratio of pleural fluid and serum of CA125 level was higher in groups A and B than in groups C, D, E. However, it is not statistically significant (Table 4). Many of the laboratory analysis of pleural fluid did not provide statistically significant information for differential diagnosis of tuberculous pleuritis except the serum and pleural fluid ADA level (p<0.05 between Group B & E) (Table 4).
Tuberculous pleuritis is thought to result from the rupture of a subpleural focus of tubercle bacilli into the pleural space and a resultant local cell-mediated immune response to mycobacterial antigens1ŌĆō3). The IS6110 insertion sequence was more sensitive than the protocol used to detect the gene coding for the 65 kD mycobacterial antigen sequences because the IS6110 is present in 10ŌĆō15 copies in each mycobacterial genome, whereas the gene coding for the 65kD antigen is probably present in only a single copy8, 16). Many studies have shown that the primer has different sensitivity and specificity for the detection of mycobacterial DNA. We used the primer set for 285 bp in IS6110, which showed generally acceptable sensitivity and specificity because the yield of PCR in tissue paraffin block might be low17, l8).
The sensitivity of the nested PCR in chronic granulomatous pleuritis with or without caseation would be increased with fresh pleural tissue and large amount of tissue. Ghossein et al. & Popper HH et al. insisted that mycobacteria might be missed in the usual 4ŌĆō6 ╬╝m section19, 20). In this study, to increase sensitivity, we used 7 slices of 7 ╬╝m thick paraffin embedded tissue in each test.
Although many of the laboratory findings of pleural fluid are used in the diagnosis of pleural effusion, none of them provides a definitive diagnosis, except pleural biopsy. Our data also showed that lots of laboratory findings of pleural fluid, including CA125, protein, glucose and lymphocyte count, did not provide definitive clues to differential diagnosis, except the ADA level between chronic granulomatous pleuritis without caseous necrosis and malignancy induced pleural effusion.
Even in patients with nonspecific or chronic inflammatory pleuritis (Group C), mycobacterial DNA was detected using nested PCR in pleural biopsy tissue. The reason why the sensitivity in Group C was quite low (25%) suggests that first, the effusion was not caused by tuberculosis even though clinically diagnosed by exclusion and a therapeutic trial; second, the pleural effusion responded to only local cell mediated immunologic reaction; third, an inadequate biopsy was performed or an incorrect process might have been done during a procedure; last, Group C has lesser content of the mycobacterial genome than Group A or B.
In this study, pleural effusion combined pulmonary infiltration which was suspected tuberculosis, was well correlated with a positive PCR reaction of mycobacterial DNA. This means that a cell-mediated immune response to a mycobacterial antigen-associated pleural effusion might have few tuberculous bacilli in the pleural tissue and resulted in a negative PCR reaction.
To avoid a false-positive diagnosis of tuberculosis, it may be necessary to quantify the amount of target DNA according to the type of specimens. Using pleural tissue is probably more specific than other specimens, such as sputum, because false positive were reported as less than 1%21). However, in control group E of this study, which was associated with malignancies, mycobacterial DNA was not detected at all. This result will be helpful to make a differential diagnosis with tuberculous pleurisy and malignant pleural effusion in elderly patients.
In this study, the pathologic findings are significantly associated with the detection rate of mycobacterial DNA. The nested PCR for Mycobacterium tuberculosis with pleural biopsy can be used as a rapid diagnostic or differential diagnostic method in not only chronic granulomatous pleuritis without caseous necrosis but also in patients with chronic or non-specific pleuritis.
Figure┬Ā1.
Polymerase chain reaction amplification of mycobacterial DNA in pleural tissues. Positive control was the H37Rv strain of M. tuberculosis (H37) and negative control was M. avium (MA). M; 100 bp DNA molecular weight marker (New England BioLabs, Inc. USA). All specimens were positive except 3rd, 7th and MA lane.
Table┬Ā1.
The sequences of primer pairs used in this study
| outer primer | IS-1 | 5ŌĆ▓-AGGCGTTGGTTCGCGAGGG-3ŌĆ▓ |
| IS-2 | 5ŌĆ▓-TGATGACGCCCTCGTTGCC-3ŌĆ▓ | |
| inner primer | IS-3 | 5ŌĆ▓-CCAACCCGCTCGGTCTCAA-3ŌĆ▓ |
| IS-4 | 5ŌĆ▓-ACCGATGGACTGGTCACCC-3ŌĆ▓ |
Table┬Ā2.
Demographic data according to pathologic groups of study population
| Group | Group A | Group B | Group C | Group D | #Group E |
|---|---|---|---|---|---|
| Pathologic Finding | CGP with caseation | CGP without caseation | Chronic or non-specific pleuritis | Congestive Heart Failure | Malignant diseases |
| Number of Patients (Male) | 7 (5) | 19 (9) | 12 (11) | 3 (3) | 11 (5) |
| Age (years) | 37.0┬▒17.2 | 37.0┬▒17.2 | 43.3┬▒15.3 | 59.3┬▒15.0 | 54.3┬▒12.0 |
| Positive Results in Nested-PCR (%) | 5 (71.4) | 7 (36.8) | 3 (15.8) | 0 | 0 |
| Clinical Diagnosis | TP* | TP* | TP** | Congestive heart failure | Malignancies* |
Table┬Ā3.
Correlation between PCR results and radiological infiltrations in tuberculous pleurisy including group A, B and C
| PCR (+) | PCR (ŌłÆ) | |
|---|---|---|
| No. of patients with RI* (%) | 13 (86.7%) | 9 (39.1%) |
| No. of patients without RI* (%) | 2 (13.3%) | 14 (60.9%) |
Table┬Ā4.
Comparison between pathologic findings and laboratory findings of pleural fluid
| Group A | Group B | Group C | Group D | Group E | |
|---|---|---|---|---|---|
| B-CA125 | 95.4┬▒41.3 | 160.7┬▒191.4 | 157.6┬▒170.8 | 351.9┬▒95.9 | 196.0┬▒174.6 |
| P-CA125 | 448.1┬▒115.9 | 468.0┬▒139.9 | 323.3┬▒146.2 | 396.5┬▒169.6 | 409.4┬▒144.8 |
| P/B CA125 | 5.8┬▒2.0 | 5.4┬▒2.8 | 3.2┬▒2.3 | 1.1 ┬▒0.2 | 3.4┬▒2.3 |
| B-ADA* | 29.5┬▒12.7 | 19.8┬▒5.9 | 29.4┬▒23.5 | 10.8┬▒4.9 | 15.6┬▒3.9 |
| P-ADA* | 61.4┬▒21.3 | 64.3┬▒16.3 | 30.9┬▒29.6 | 4.1┬▒2.8 | 19.9┬▒7.3 |
| P/B-ADA* | 2.3┬▒1.1 | 3.5┬▒1.2 | 1.4┬▒1.3 | 0.4┬▒0.2 | 1.3┬▒0.3 |
| B-Protein | 6.8┬▒0.9 | 7.0┬▒1.0 | 6.9┬▒1.0 | 6.9┬▒0.2 | 7.0┬▒0.5 |
| P-Protein | 4.9┬▒1.6 | 6.1┬▒2.7 | 6.3┬▒2.4 | 3.2┬▒1.1 | 5.1┬▒1.8 |
| P/B-Protein | 0.7┬▒0.2 | 0.9┬▒0.3 | 0.9┬▒0.3 | 0.5┬▒0.2 | 0.7┬▒0.2 |
| RBC | 2486┬▒1518 | 8327┬▒2143 | 12683┬▒2584 | 5436┬▒2624 | 107484┬▒42183 |
REFERENCES
2. Falk A. Tuberculous pleurisy with effusion: Diagnosis and results of chemotherapy. Postgrad Med 38:631ŌĆō6351965.


3. Levine H, Metzger W, Lacera D. Diagnosis of tuberculous pleurisy by culture of pleural biopsy specimen. Arch Intern Med 126:269ŌĆō2711970.


5. Peterson EM, Floyd RC, Nakasone A, Fridly G, Maza LM. Direct identification of Mycobacterium tuberculosis Mycobacterium avium and Mycobacterium intercellulare from amplified primary cultures in BACTEC media using DNA probes. J Clin Microbiol 27:1543ŌĆō15471989.



6. Collins DM, Lisle GW. DNA restriction endonuclease analysis of Mycobacterium tuberculosis and Mycobacterium bovis BCG. J Gen Microbiol 130:1019ŌĆō10231984.


7. Kim HJ, Kim YW, Han SK, Shim YS, Kim KY, Han YC. Identification of Mycobacterium tuberculosis in pleural effusion by polymerase chain reaction. Tubercul Respir Dis 40:509ŌĆō5181993.

8. Lee BW, Tan JAMA, Wong SC, Tan CB, Yap HK, Low PS, Chia JN, Tay JSH. DNA amplification by the polymerase chain reaction for the rapid diagnosis of tuberculous meningitis Comparison of protocols involving three mycobacterial DNA sequences IS6110, 65 kDa antigen, and MPB64. J Neurol Sci 123:173ŌĆō1791994.


9. Patel R, Kvach J, Mounts P. Isolation and restriction endonuclease analysis of mycobacterial DNA. J Gen Microbiol 132:541ŌĆō5511986.


10. Brisson-Noel A, Gicquel B, Lecossier D, Levy Frebault V, Nassif X, Hance AJ. Rapid diagnosis of tuberculosis by amplification of mycobacterial DNA in clinical samples. Lancet 4:1069ŌĆō10711989.

11. Thieery D, Brisson-Noel A, Levy-Frebault V, Neguyen S, Guesdon JL, Gicquel B. Characterization of a Mycobacterium tuberculosis insertion sequence, IS6110, and its application in diagnosis. J Clin Microbiol 28:2668ŌĆō26711990.



12. Lassence A, Lecossier D, Pierre C, Cadranel J, Stern M, Hance AJ. Detection of mycobacterial DNA in pleural fluid from patients with tuberculous pleurisy by means of the polymerase chain reaction comparison of two protocols. Thorax 47:265ŌĆō2691992.



13. Pierre C, Lecossier D, Boussougant Y. Use of a reamplification protocol improves sensitivity of detection of mycobacterium tuberculosis in clinical samples by amplification of DNA. J Clin Microbiol 29:712ŌĆō7171991.



14. Saiki RK, Genfand DH, Stoffel SJ, Higuchi R, Horn GT, Mullis KB, Ehrich HA. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science 239:487ŌĆō4911988.


15. Hance AJ, Grandchamp B, Lavy-frebault V. Detection and identification of mycobacteria by amplification of mycobacterial DNA. Mol Microbiol 3:843ŌĆō8491989.


16. Walker DA, Taylor IK, Mitchell DM, Shaw RJ. Comparison of polymerase chain reaction amplification of two mycobacterial DNA sequences IS6110 and 65KDa antigen gene in the diagnosis of tuberculosis. Thorax 47:690ŌĆō6941992.



17. Kim SJ, Park YK, Cho SH, Shim MS. Primer-directed amplification of Mycobacterium tuberculosis DNA in clinical specimens I Primers and reaction conditions. J Korean Soc Microbiol 27:35ŌĆō441992.
18. Eisenach KD, Stifford MD, Cave MD, Bates JH, Crawford JT. Detection of mycobacterium tuberculosis in sputum samples using a polymerase chain reaction. Am Rev Resp Dis 144:1160ŌĆō11631991.


19. Ghossein RA, Ross DG, Salomon RN, Rabson AR. Rapid detection and species identification of mycobacteria in paraffin-embedded tissues by polymerase chain reaction. Diagn Mol Pathol 1:185ŌĆō1911992.





 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Download Citation
Download Citation Print
Print



