The effects of transferring tumor suppressor gene p16INK4A to p16INK4A-deleted cancer cells*
Article information
Abstract
Objectives
p16 is known to be an important tumor suppressor gene and is also called MTS1(multiple tumor suppressive gene 1). Especially in the case of non-small cell lung cancer, it was not expressed in more than 70% of cell lines examined. To determine changes in cell-cycle related proteins and the tumorigenic effect, we, therefore, transfected p16INK4A gene into lung cancer cell lines.
Methods
We transfected p16INK4A gene into lung cancer cell lines which do not express p16 protein. We evaluated the effect by clonogenic assay and observed the changes of cell-cycle related proteins.
Results
The newly-expressed p16 formed a complex with cdk4, and phosphorylated pRB was decreased, although cyclin D1 and pRB:cyclin D1 complex were unchanged. Clonogenic assay after selection with G4T8 showed that, in the cell lines transfected with p16, tumorigenicity was significantly less than in the control.
Conclusion
These results suggest that the p16INK4A gene can be a candidate for gene therapy in cases of NSCLC in which p16INK4A gene is inactivated.
INTRODUCTION
It is clear that in human cancer, including lung cancer, deregulation of cell cycle progression is a hallmark of neoplastic transformation and that genes involved in G1/S transition of the cell cycle are especially frequent targets for mutations. A cell-cycle related protein, the recently identified p16INK4A gene product, plays an important role in the negative regulation of the kinase activity of cyclin dependent kinase(cdk) enzymes1). p16INK4A gene is, thus, an important tumor suppressor gene and is also called MTS1(multiple tumor suppressive gene 1)2, 3). In various malignancies, reports of alterations in the p16INK4A gene are much more common than those involving any other oncogene or tumor-suppressive gene ever identified2, 4, 5) It was particularly noticeable that there was no p16 expression in more than 70% of cell lines examined in the case of non-small cell lung cancer. It is speculated that p16INK4A gene could exert a key role in the development of non-small cell lung cancer6). The purpose of this study was to investigate whether p16INK4A gene is a suitable candidate for gene therapy in cases of non-small cell lung cancer. After the extraction of total RNA from normal fibroblast cell lines and subsequent reverse transcriptase and polymerase chain reaction, amplified p16INK4A was subcloned into eukaryotic expression plasmid vector, pRC-CMV. Using lipofectin, the constructed pRC-CMV-p16 was transfected into NSCLC cell lines NCI-H441 and NCI-H157. After the extraction of proteins from cell lysates, the changes in G1 cell-cycle related proteins were investigated with Western blot analysis and immunoprecipitation and the tumor suppressive effect was observed by clonogenic assay.
MATERIALS AND METHODS
Cell lines
Non-small cell lung cancer cell lines NCI-H441, NCI-H157 and NCI-H2009 were kindly donated by Frederic Kaye (National Cancer Institute, U.S.A.) and were grown in RPMI media supplemented with 10% fetal bovine serum and antibiotics. They were maintained at 37°C in a 5% CO2 incubator and the media was changed twice a week.
Construction of p16 expression plasmid
p16 expression plasmid(pRC-CMV-p16) was kindly donated by Dr. Gregory Otterson (National Cancer Institute, U.S.A.). For the construction of p16 expression plasmid, RT-PCR was performed using mRNA from NIH3T3 fibroblast cell line (sense primer; 5′ AAG-CTT-ATG-GAG-CCT-TCG-GCT 3′, anti-sense primer; 5′ TTC-GAA-TCA-ATC-GGG-GAT-GTC 3′), and p16 cDNA was made. The lengh of p16 cDNA was 8 amino acids shorter than the natural full length. The PCR product was cloned into pRC-CMV plasmid, and the sequence was verified by sequencing. Using the cesium chloride method, an adequate quantity of pRC-CMV-p16 plasmid was obtained.
Transfection
Lung cancer cell lines, NCI-H441 and NCI-H157, were transfected via lipofectin, as described by the manufacturer (Life Technologies). Ten micrograms of pRC-CMV-p16 or control pRC-CMV plasmid were transfected into 100-mm-diameter subconfluent plates.
Western blot analysis and Immunoprecipitation
Cellular protein was extracted 60 hours after transfection by lysing cells with lysis buffer(50 mM Tris-HCl, pH 7.5/250 mM NaCl/5 mM EDTA/0.1% Nonidet P-40(NP-40)/50 mM NaF/1 mM phenylmethylsulfonyl fluoride(PMSF)) containing protease inhibitors(10 ug/ml leupeptin and 50 ug/ml aprotinin). The lysate was incubated for 20 min on ice and was centrifuged(14,000 X g for 20 min); the supernatant was frozen at −70°C until used.
For Western blot analysis, 200 ug of cellular proteins were electroblotted to nitrocellulose paper after separation on 7.5 – 15 % SDS-PAGE and then allowed to incubate overnight at 4°C with a 1:2000 dilution of primary antibody(anti-human RB, p16, cdk4 or cyclin D1) in PBS containing 5% powdered milk and 1% bovone serum albumin. Conjugation with secondary antibodies and visualization was performed using an ECL kit(Amersham) as recommended by the manufacturer.
For immunoprecipitation, 500 ug of cellular proteins were immunoprecipitated with 10 ug of anti-human CDK4 or anti-human cyclin D1 antibodies. The immunoprecipitated proteins, separated by protein A-sepharose beads, were immunoblotted with anti-human p16 or anti-human RB antibodies.
Antibodies used for Western blot analysis and immunoprecipitation were mouse monoclonal anti-human RB, mouse monoclonal anti-human cyclin D1, rabbit polyclonal CDK4 and rabbit polyclonal anti-human p16(PharMingen, San Diego).
Clonogenic assay
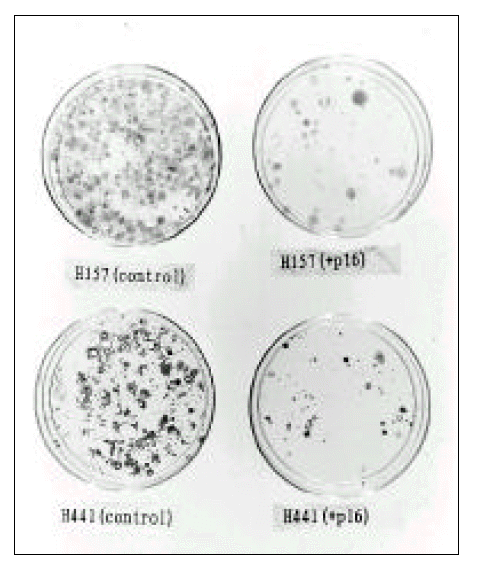
After transfection, transfected cells were selected using G418(150 ug/ml) containing media. When visible colonies were seen, the plates were fixed with methanol and stained with crystal violet, and the numbers of colonies were compared.
RESULTS
Expression of p16 protein in transfected cells
Transfection of pRC-CMV-p16 to p16(−) NCI-H441 cells resulted in p16 expression equal to that seen in non-transfected NCI-H2009, which expresses p16 normally(Fig. 1). Control NCI-H441 cells did not express p16 protein(Fig. 1), and this indicates that in non-small cell lung cancer cell lines, the pRC-CMV-p16 expression vector is very useful for transfection.

Western blot analysis of p16 protein expressed in NCI-H2009, NCI-H441 and NCI-H441-p16 cell line transfected with pRC-CMV-p16. Lane 1 (NCI-H2009) shows a positive signal with polyclonal rabbit anti-human p16, but lane 2 (NCI-H441) is negative. Lane 3 (NCI-H441-p16) shows positive signal in slightly lower position because of shorter lengh of the construct.
p16:cdk4 complex formation of expressed p16 after transfection with pRC-CMV-p16
p16 protein, expressed in NCI-H441 cells after transfection with pRC-CMV-p16, formed a complex with cdk4(Fig. 2). p16 is known to inhibit cdk4 by forming a complex with it and inhibiting the phosphorylation of cdk4 substrate. The fact that p16 protein expressed after transfection formed this complex indicates that p16 functioned normally.
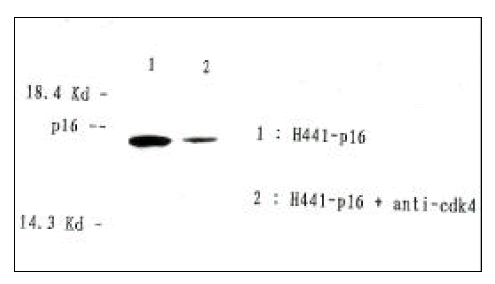
Identification of p16:cdk4 complex formation in p16(−) NCI-H441 cell line after transfection with pRC-CMV-p16. Extracted protein solution was immunoprecipitated with polyclonal rabbit anti-human cdk4 specific antibody and protein A-sepharose beads. After SDS-PAGE of separated proteins by 15% gel, Western blot analysis was performed with polyclonal rabbit anti-human p16 as the primary antibody. Lane 2 (NIH-H441-p16 + anti-cdk4) shows a positive signal in the same position as p16 in lane 1.
Dephosphorylation of pRB protein after transfection with pRC-CMV-p16
To determine the effect of p16 protein expressed after transfection, changes in pRB protein in NCI-H441 cells were investigated. As expected, the amount of phosphorylated pRB protein was less than the amount of phosphorylated pRB protein in control cells (Fig. 3). This means that p16 protein expressed after transfection of pRC-CMV-p16 inhibited the phosphorylation of pRB protein by cdk4.
Changes in cyclin D1 and cyclin D1:cdk4 complex after transfection with pRC-CMV-p16
Because p16 protein expressed after transfection inhibited the phosphorylation of pRB, we investigated changes in cyclin D1 and cyclin D1:cdk4 complex, known to be involved in the phosphorylation of pRB. No significant change was apparent in cyclin D1. However, the change in cyclin D1:cdk4 complex could not be determined because baseline cyclin D1:cdk4 complex band was too faint in our experiment(Fig. 4).

Identification of cyclin D:cdk4 complex formation in NCI-H441 and NCI-H441-p16 cell lines transfected with pRC-CMV-p16. Extracted protein lysate was immunoprecipitated with polyclonal rabbit anti-human cdk4 specific antibody and protein A-sepharose beads. After SDS-PAGE of separated proteins by 12.5% gel, Western blot analysis was performed with monoclonal mouse anti-human cyclin D1 as the primary antibody. At the cyclin D1 position, both lane 3 (NCI-H441 + anti-cdk4) and lane 4 (NCI-H441-p16 + anti-cdk4) show very faint signals, in contrast to both lane 1 (NCI-H441) and lane 2 (NCI-H441-p16). Cyclin D1 expression was, therefore, the same before and after transfection with pRC-CMV-p16. However, the change in cyclin D1:cdk4 complex formation could not be determined in this experiment.
Change in the pRB:cyclin D1 complex after transfection with pRC-CMV-p16
In vivo, cyclin D1 is known to form a complex with pRB, thus affecting the phosphorylation of pRB. After transfection, no change in the pRB:cyclin D1 complex was noted (Fig. 5), suggesting that p16 does not function through change in the pRB:cyclin D1 complex.

Identification of pRb:cyclin D1 complex formation in NCI-H441 and NCI-H441-p16 cell lines transfected with pRC-CMV-p16. Extracted protein lysate was immunoprecipitated with monoclonal mouse anti-human cyclin D1 specific antibody and rabbit anti-mouse IgG-protein A-sepharose beads. After SDS-PAGE of separated proteins by 7.5% gel, Western blot analysis was performed with monoclonal mouse anti-human pRb as the primary antibody. At the pRb position, both lane 2 (NCI-H441 + anti-cyclin D1) and lane 4 (NCI-H441-p16 + anti-cyclin D1) show positive signals, as do lane 1 (NCI-H441) and lane 3 (NCI-H441-p16). This indicates that pRb:cyclin D1 complex formation was the same before and after transfection with pRC-CMV-p16.
Tumor suppressive effect of p16
The colony-forming ability of NCI-H441 and NCI-H157 cell lines after transfection with pRC-CMV-p16 was compared with that of control cell lines transfected with pRC-CMV. Significantly fewer colonies were observed in plates transfected with pRC-CMV-p16 than in those transfected with pRC-CMV(Fig. 6), and this suggests that p16 may have a tumor suppressive effect on non-small cell lung cancer cell lines which do not express p16.
DISCUSSION
A variety of evidence suggests that p16 is a tumor suppressor gene. p16INK4A is located in chromosome 9p21, where deletions have frequently been found in carcinomas of lung, kidney, ovary, pancreas, breast, bladder, head and neck and esophagus and in several other tumors, such as glioma, melanoma, leukemia and osteogenic sarcoma2, 3, 7–9). In addition, homozygous deletions or mutations of the p16INK4A gene are also frequently observed in cell lines from these tumors4, 10). Because deletion or mutation of p16INK4A is significantly less frequent in primary cancer tissues than in cancer cell lines, it has been suspected, however, that p16INK4A may not be a true tumor suppressor gene5, 11). In cases of non-small cell lung cancer, about 70% of cell lines do not express p16 protein6), but mutation of the p16INK4A gene is not frequent in primary tissues12).
To confirm the tumor suppressive effect and the mechanisms of p16INK4A gene in non-small cell lung cancer, we transfected p16INK4A gene into cell lines of this cancer via lipofectin. p16(−) NCI-H441 cell line transfected with pRC-CMV-p16 showed the formation of p16:cdk4 complex and decreased phosphorylated Rb protein, while the control cell line did not. These results are consistent with those of Sandig et al., who studied the expression of pRB in a hepatocellular carcinoma cell line13). pRB has been shown to play a protective role in cell apoptosis14). When its levels are sufficiently reduced, this role can no longer be maintained, and this may explain part of the tumor suppressor function of p16. Clonogenic assay demonstrated that the extent of colony formation was significantly less in p16(−) NCI-H441 and NCI-H157 cell lines transfected with pRC-CMV-p16 than in the control cell line. This study shows that the expression of p16 protein in gene-transfected NSCLC cell lines, in which p16 is absent, leads to p16:cdk4 complex formation and a subsequent decrease in phosphorylated pRb protein levels, and leads ultimately to tumor suppressive effects.
Using pRC/CMV plasmid vector, Okamoto et al. showed that when p16INK4A gene transfer was involved, colony formation by esophageal cancer cell lines decreased12). Arap et al. demonstrated the growth suppressive effect of p16INK4A gene in p16(−) glioma cell lines15), and this finding is in agreement with those of other reports.16, 17) Though this effect was not observed in glioma cell lines which express wild-type p16 protein15). It has been suggested that CDK4 or cyclinD may be amplified or overexpressed18, 19), or a G1/S cell cycle pathway such as p53-p21WAF1 may be inactivated in cases of glioma with wild-type p16 expression15). This suggests that, for the general use of gene therapy for the treatment of cancer, combination gene therapy involving p53 and p16, for example, may be necessary.
Tumor suppressor gene therapy requires efficient delivery of therapeutic genes and a high level of expression of the newly transduced gene in the tumor. The adenovirus is an efficient expression vector that can be produced in high titers20). Its expression time in the host cell is short, and it is thus suitable for use in gene replacement strategies involved in cancer treatment. For the clinical trial of p16 in cancer gene therapy, adenovirus vector is one of the suitable candidates. A few in vitro and animal studies on p16 in the adenovirus vector have been published recently16, 17, 21).
It was difficult to obtain cell lines in which the expression of p16 protein after transfection of p16INK4A gene in NSCLC cell lines was stable. Because p16-transfected cells which overexpress p16 do not usually survive, we could not obtain data relating to flow cytometric cell cycle analysis or quantitative cytotoxicity tests. This difficuty is a common problem in the study of tumor suppressor genes and, to overcome it, an inducible promotor is usually used in the cloning of expression vectors.
In this study, we confirmed in vitro that after transfection of p16INK4A gene to a cell line which has wild-type pRB but does not express p16, p16 formed a complex with cdk4, phosphorylation of pRB decreased, and there was decreased colony formation. This suggests that the p16INK4A gene can be a candidate for gene therapy in cases of NSCLC in which p16INK4A gene is inactivated.