Genetic susceptibility to post-endoscopic retrograde cholangiopancreatography pancreatitis identified in propensity score-matched analysis
Article information
Abstract
Background/Aims
A previous history of post-endoscopic retrograde cholangiopancreatography (ERCP) pancreatitis (PEP) is a risk factor for PEP, suggesting that there may be a genetic predisposition to PEP. However, nothing is known about this yet. The aim of this study was to identify genetic variations associated with PEP.
Methods
A cohort of high-risk PEP patients was queried from December 2016 to January 2019. For each PEP case, two propensity score-matched controls were selected. Whole exome sequencing was performed using blood samples. Genetic variants reported to be related to pancreatitis were identified. To discover genetic variants that predispose to PEP, a logistic regression analysis with clinical adjustment was performed. Gene-wise analyses were also conducted.
Results
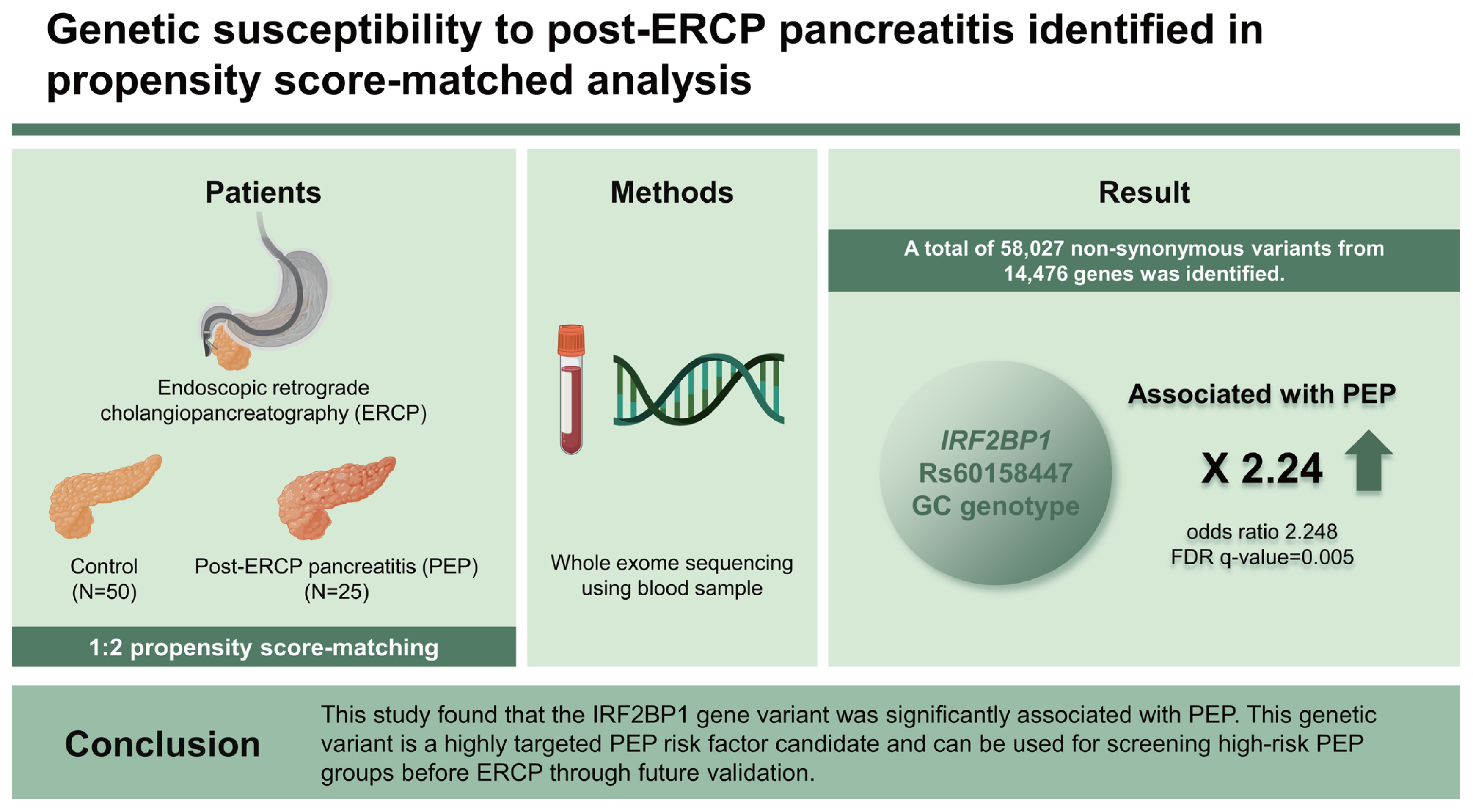
Totals of 25 PEP patients and 50 matched controls were enrolled. Among the genetic variants reported to be associated with pancreatitis, only CASR rs1042636 was identified, and it showed no significant difference between the case and control groups. A total of 54,269 non-synonymous variants from 14,313 genes was identified. Logistic regression analysis of these variants showed that the IRF2BP1 rs60158447 GC genotype was significantly associated with the occurrence of PEP (odds ratio 2.248, FDR q value = 0.005). Gene-wise analyses did not show any significant results.
Conclusions
This study found that the IRF2BP1 gene variant was significantly associated with PEP. This genetic variant is a highly targeted PEP risk factor candidate and can be used for screening high-risk PEP groups before ERCP through future validation. (ClinicalTrials.gov no. NCT02928718)
INTRODUCTION
Post-endoscopic retrograde cholangiopancreatography (ERCP) pancreatitis (PEP) is one of the most common complications of ERCP, occurring in 2–9% of patients undergoing ERCP [1]. Most PEPs have a mild clinical course, but rarely have a fatal clinical course, resulting in an overall mortality rate of about 0.7% [2]. Accordingly, many studies on risk factors and prevention of PEP have been conducted, and the previous history of PEP is one of the well-known risk factors for PEP [3,4]. In a systematic review of risk factors for PEP, Chen et al. reported that patients with prior PEP had a 2.23 times higher incidence of PEP than controls [5]. As such, previous PEP history is a risk factor for its occurrence, suggesting a predisposition to PEP, which may be genetic in nature. However, few studies have explored the genetic susceptibility associated with PEP. One study evaluated N34S mutation in the SPINK1 gene, which is associated with development of pancreatitis, in 30 patients with PEP using polymerase chain reaction (PCR) with restriction fragment length polymorphism; no significant results were obtained [6]. There was also a case-control study of the association between genetic variants in PRSS1-PRSS2 and MORC4, which are related to alcoholic pancreatitis, and PEP, but did not find a significant association [7]. Both previous studies only assessed associations between a few genetic variants and PEP and failed to confirm associations. Thus, there is a need to examine the association between more diverse genetic variants and PEP. In recent years, as next-generation sequencing (NGS) has become more common and less expensive, gene variation analysis has become easier [8]. Therefore, the present study, using NGS, aimed to evaluate whether various known genetic mutations related to pancreatitis are related to the occurrence of PEP, and to explore such novel genetic variants.
METHODS
Patients and data collection
This study prospectively enrolled patients from three academic medical centers who were at high risk for PEP and underwent ERCP from December 2016 to January 2019. High-risk patients for PEP were defined as those who had at least one of the following conditions [9]: 1) clinically suspected sphincter of Oddi dysfunction (SOD) (defined the same as the previous study [9]), 2) history of PEP, 3) pancreatic sphincterotomy, 4) precut sphincterotomy (if performed due to failure of standard cannulation technique), 5) difficult cannulation (cannulation taking more than 10 minutes or more than eight cannulation attempts), 6) balloon dilatation of sphincter without sphincterotomy, 7) endoscopic ampullectomy, and 8) two or more of the following conditions (female sex, younger than 50 years, history of recurrent pancreatitis, at least three injections of contrast media into the pancreatic duct with at least one into the pancreatic tail, pancreatic acinar opacification due to over-injection of contrast media into the pancreatic duct, and brush cytology examination of the pancreatic duct). Patients with at least one of the following conditions were excluded: 1) younger than 18 years, 2) acute pancreatitis within 72 hours prior to ERCP, 3) pregnant or breast-feeding, 4) patient refusal to participate in the study, 5) contraindication of ERCP, 6) patients with prior endoscopic sphincterotomy (EST) who were expected to undergo only bile duct-related procedures, such as replacement or removal of a biliary stent, 7) chronic pancreatitis, 8) history of gastrectomy with Billroth II or Roux-en Y anastomosis, and 9) pancreatic or distal bile duct cancer. The study also excluded patients with asymptomatic hyperamylasemia and missing blood test results before performing propensity score (PS) matching between PEP patients and controls.
Demographic and clinical data including age, sex, main indications for ERCP, history of PEP, clinically suspected SOD, and abdominal pain assessed by a numeric rating scale at four hours and between 12 and 24 hours after ERCP were collected. Laboratory values such as white blood cell count and liver function tests were measured within 24 hours before ERCP, and serum amylase and lipase were measured at four hours and between 12 and 24 hours after ERCP. Endoscopic data including periampullary diverticulum, biliary sphincterotomy, pancreatic sphincterotomy, precut sphincterotomy, endoscopic papillary balloon dilatation (EPBD), pancreatogram, pancreatic duct brush cytology, cannulation success, difficult cannulation, endoscopic ampullectomy, pancreatic stent insertion, biliary stent insertion, and ERCP procedure time were also collected. This study was registered at clinicaltrials.gov (NCT02928718), complied with the Declaration of Helsinki, and was approved by the institutional review board of each institution (Seoul National University Hospital IRB No. 1507-124-689, Dongguk University Ilsan Hospital IRB No. 2017-11-017, and Seoul National University Boramae Medical Center IRB No. 16-2016-135). Written informed consent was obtained from all participants. Most patients at high risk of potential PEP, including patients scheduled for endoscopic ampullectomy, were enrolled before ERCP, and the remaining patients, including those whose PEP risk was difficult to predict before ERCP, were enrolled after ERCP.
Study outcomes and definition
The study outcome of interest was a genetic variant significantly associated with development of PEP. PEP was defined as a case in which serum amylase or lipase was elevated to more than three times the upper limit of normal, and new onset upper abdominal pain persisted for more than 24 hours after ERCP [10]. PEP severity was defined according to the consensus criteria: mild (hospitalization for 2–3 days), moderate (hospitalization for 4–10 days), and severe (hospitalization for more than 10 days) [10].
Sample acquisition, DNA preparation and whole exome sequencing
After informed consent was obtained, four milliliters of whole blood were drawn from each participants and collected in EDTA-containing tubes. Samples for NGS were collected at the same time as blood draws for laboratory measurements within 24 hours after ERCP, and immediately frozen and stored at −20°C until DNA extraction. Genomic DNA (gDNA) was extracted using a DNeasy Blood & Tissue Kit (Qiagen, Germantown, MD, USA; Cat No. 69504), according to the manufacturer’s instructions. The extracted gDNA was quantified using Quant-iT PicoGreen dsDNA Assay kit (ThermoFisher Scientific, Waltham, MA, USA; Cat No. P11495). To assess the integrity of the gDNA, samples were run on the TapeStation4200 gDNA ScreenTape (Agilent Technologies, Waldbronn, Germany). Only high-quality gDNA with a DNA integrity number greater than 7.0 was used for library construction. For generation of standard exome capture libraries, the Agilent SureSelect Target Enrichment protocol for Illumina paired-end sequencing library was used together with 1 μg of input gDNA. In all cases, the SureSelect Human All Exon V6 probe set was used. Then 1 μg of each gDNA diluted in EB Buffer was sheared to a target peak size of 150–200 bp using the Covaris LE220 focused-ultrasonicator (Covaris, Woburn, MA, USA) according to the manufacturer’s recommendations. The 8 microTUBE Strip was loaded into the tube holder of the ultrasonicator, and the DNA was sheared using the following settings: mode, frequency sweeping; duty cycle, 10%; intensity, 5; cycles per burst, 200; duration, 60 s × 6 cycles; temperature, 4–7°C. The fragmented DNA was repaired, an ‘A’ was ligated to the 3′ end, and SureSelect adapters were ligated to the fragments. Once ligation had been assessed, the adapter ligated product was PCR amplified. The final purified product was then quantified using the TapeStation4200 D1000 ScreenTape (Agilent Technologies). For exome capture, 250 ng of DNA library was mixed with hybridization buffers, blocking mixes, RNase block, and 5 μL of SureSelect all exon capture library, according to the standard Agilent SureSelect Target Enrichment protocol. Hybridization to the capture baits was conducted at 65°C using a heated thermal cycler lid option at 105°C for 24 hours on a PCR machine. The captured DNA was washed and amplified. The final purified product was quantified using qPCR according to the qPCR Quantification Protocol Guide (KAPA Library Quantificatoin kits for Illumina Sequecing platforms) and qualified using the TapeStation4200 D1000 ScreenTape (Agilent Technologies). Then the NovaSeq6000 platform (Illumina, San Diego, CA, USA) was used to sequence.
Analysis of genetic variants known to be associated with pancreatitis
The researchers confirmed whether the following genetic mutations, known to be related to pancreatitis in previous reports, were present in this study’s subjects: PRSS1 R122H [11], N29I [11], A16V [12]; PRSS2 G191R [13]; SPINK1 N34S [14]; CFTR F508del [15], R75Q [16]; CTRC R254W [17], K247_R254del [17]; CASR R990G [18], R896H [19]; CPA1 N256K [20].
Genetic variant calling
The base calling files were converted to FASTQ files by an Illumina package. The sequencing quality was checked using FastQC [21]. Paired-end sequence files were mapped to the human reference genome version 19 (hg19) from UCSC using bwa-0.7.17 [22], specifically the bwa-mem algorithm. The mapping result was generated in a BAM format. PCR duplicates were marked and removed using the MarkDuplicate tool from Picard-2.18.2 [23]. After removing duplicates, BAM files were realigned for small indels and recalibrated with Base Quality Score Recalibration (BQSR) tool from the Genome Analysis ToolKit (GATK) 4.0.5.1 [24]. Based on the BAM files pre-processed, variant calling for each sample was conducted with HaplotypeCaller gvcf mode from GATK. For variant-level quality control, variants were filtered with VariantFiltration from GATK using default options. Analysis-ready files were generated in VCF format. All VCF files were annotated using SNPEff (released 2021-03-09) [25]. The sequence alignment and the quality of single nucleotide variants (SNVs) were manually examined using IGV 2.8.9 [26] to exclude false-positive SNVs (Supplementary Fig. 1).
Statistical analysis
Continuous data were presented as a mean ± standard deviation and categorical data were presented as a number with a percent. Comparisons between groups were performed using Student’s t-test for continuous variables and a chi-square test or Fisher’s exact test for categorical variables. PS matching was performed on clinical factors that differed between PEP and the control groups by a p value less than 0.1 to reduce the difference in clinical factors that may affect the occurrence of PEP. With this PS matching, two matched controls were selected for each PEP case. Associations between the occurrence of PEP and filtered nonsynonymous variants were analyzed in three ways: 1) At the variant level, the logistic regression test was performed for the following three modes of inheritance: additive, dominant, and recessive. The logistic regression test was adjusted with clinical factors including sex, age, pancreatic EST, precut EST, EPBD, pancreatogram, cannulation success, difficult cannulation, clinically suspected SOD, history of PEP, endoscopic ampullectomy, pancreatic stent insertion, and ERCP procedure time. 2) Regarding gene level, the gene-wise variant burden (GVB) score [27] of each gene was calculated. To compare GVB scores between PEP cases and controls, Student’s t-test was conducted. 3) To assess the effects of rare variants, an optimal sequence kernel association test (SKAT-O) [28] was performed with rare variant (1000 genome population allele frequency < 0.05) by gene [29]. The Benjamini-Hochberg multiple testing correction was performed for all three analyses to estimate the false discovery rate (FDR). A p value < 0.05 and an FDR q value < 0.05 were considered statistically significant. All statistical analyses were performed using IBM SPSS Statistics software version 24.0 (IBM Corp., Armonk, NY, USA) and R 4.1.0 software (The R Foundation for Statistical Computing, Vienna, Austria).
RESULTS
Patient characteristics
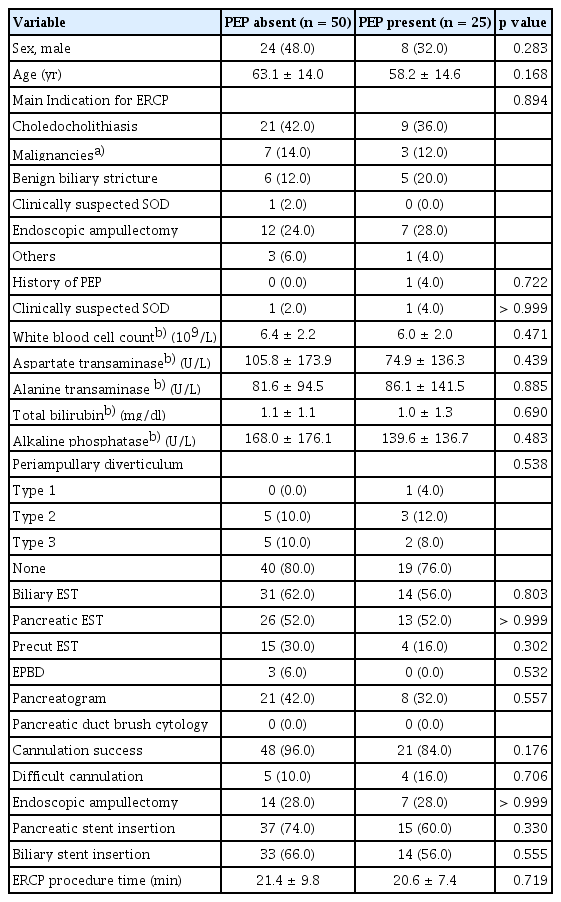
During the study period, 5,836 patients underwent ERCPs. Among these patients, 138 were screened and provided informed consent. From this group, 110 patients at high risk for PEP were selected, after excluding one patient who withdrew consent, one patient diagnosed with pancreatic cancer, 17 patients with asymptomatic hyperamylasemia, and nine patients with missing blood tests. Of these, 25 were PEP patients and 85 were controls, and their baseline characteristics are summarized in Table 1. Compared to the control group, the PEP group had a larger proportion of females and significantly lower mean serum total bilirubin and alkaline phosphatase values in blood tests performed within 24 hours prior to ERCP. Although not statistically significant, the PEP group showed a tendency for lower cannulation success rate and pancreatic stent insertion rate compared to the control group. PS matching was performed on the factors that showed differences between patients and controls, and a total of 75 subjects, consisting of 25 PEP patients and 50 controls, was finally selected for analysis (Fig. 1). Their baseline characteristics are summarized in Table 2, and no clinical factors showed a significant difference between the two groups selected through PS matching.

Study flow chart. PEP, post-endoscopic retrograde cholangiopancreatography pancreatitis; PS, propensity score.
Genetic variants known to be associated with pancreatitis in patients with PEP
The CASR R990G (rs1042636) variant was identified in 19 of 25 PEP patients (four patients with homozygous variants and 15 patients with heterozygous variants, variant allele frequency [VAF] 0.46). However, there was no significant difference in the frequency of CASR R990G variant between the PEP patients group and the control group (p = 0.568) (Table 3). Other than CASR R990G, genetic variants associated with pancreatitis investigated in this study were not identified.
Genetic variants associated with PEP
In analysis-ready VCF files, a total of 390,793 variants was called in 24,981 genes. Among these variants, after excluding 336,254 synonymous variants, 54,269 nonsynonymous variants in 14,313 genes were analyzed. The logistic regression analysis with adjustment of clinical factors showed that only one variant of the IRF2BP1 gene was significantly associated with the occurrence of PEP. Patients with the IRF2BP1 rs60158447 GC genotype (odds ratio [OR] 2.248, 95% confidence interval [CI] 1.688–2.994; FDR q value = 0.005) had a significantly higher occurrence of PEP than patients with the IRF2BP1 rs60158447 GG genotype in the dominant genetic model. No one had the homozygous CC genotype for IRF2BP1 rs60158447. Eight of 25 patients with PEP had the IRF2BP1 rs60158447 GC genotype, whereas the control group had no IRF2BP1 rs60158447 GC genotype (Table 4).
There were two genetic variants that, although not statistically significant, tended to have protective effects on PEP in the dominant genetic model. One was the rs1824152 genotype with the C allele in the CPAMD8 gene (OR 0.028, 95% CI 0.405–0.700, FDR q value = 0.074), and the other was the rs11703226 genotype with the C allele in the TUBGCP6 gene (OR 0.115, 95% CI 0.525–0.772, FDR q value = 0.074) (Table 4).
Among the 25 cases of PEP, there were 8, 15, and 2 cases of mild, moderate, and severe PEP, respectively. There was no genetic variant significantly associated with the severity of PEP.
There were no significant results in GVB score analysis or the SKAT-O test (Fig. 2).

Schematic diagram of genetic variant analysis. The diagram shows how genetic variants were analyzed. FDR, false discovery rate; GVB, gene-wise variant burden; SKAT-O, optimal sequence kernel association test; VCF, variant call format. a)Adjusted clinical variables: sex, age, pancreatic sphincterotomy, precut sphincterotomy, endoscopic papillary balloon dilatation, pancreatogram, cannulation success, difficult cannulation, clinically suspected sphincter of Oddi dysfunction, history of post-endoscopic retrograde cholangiopancreatography pancreatitis, endoscopic ampullectomy, pancreatic stent insertion, and endoscopic retrograde cholangiopancreatography procedure time.
DISCUSSION
Although various studies have been conducted on the risk factors for PEP, a common complication of ERCP, little is known about the genetic susceptibility of PEP. Therefore, in this study, we aimed to explore the genetic variants related to PEP and performed genetic variant analysis using PS-matched controls to adjust various clinical factors including ERCP procedure-related issues known as risk factors for PEP. This study found that rs60158447 in the IRF2BP1 gene was associated with the occurrence of PEP. To our knowledge, this is the first study to suggest a genetic variant associated with PEP. The IRF2BP1 rs60158447 variant shows 3% VAF in the entire 1000 genome database and 8.1% in East Asians [29]. In the PEP case group of this study, the VAF of IRF2BP1 rs60158447 was 16%, about twice the value found in the East Asian data, and the control group had a VAF of 0%, showing a significant difference between the two groups. IRF2BP1 is the protein that acts as a transcriptional co-repressor in an interferon regulatory factor-2 (IRF2)-dependent manner [30]. IRF2BP1, a member of the IRF2BP family of transcriptional regulators, contributes to the modulation of apoptosis in breast cancer cells and has been reported to play an important role in epidermal growth factor receptor signaling [31,32]. There have been no reports of a direct association between IRF2BP1 and pancreatitis. However, association between IRF2 and acute pancreatitis was reported by Mashima et al. [33]. They reported in a mouse model that IRF2 plays an important role in the process of exocytosis in the pancreas and is involved in the early phase of acute pancreatitis. They identified an early feature of acute pancreatitis in IRF2 knockout mice, where zymogen granules containing digestive enzymes were not released through the apical pole of pancreatic acinar cells due to poor exocytosis [33,34]. Acute pancreatitis is known to occur when the apical secretion of acinar cells is reduced, followed by pathogenic zymogen activation and increased basal secretion by stimuli such as alcohol, pancreatic duct obstruction by biliary stone, ERCP procedure, etc. [34]. Therefore, if the rs60158447 in the IRF2BP1 gene identified in this study acts in the direction of downregulating IRF2, the apical secretion of the zymogen granule is suppressed, making it more susceptible to pancreatitis, suggesting an increased risk of PEP. Further studies are needed on the relationship between the rs60158447 in the IRF2BP1 gene and IRF2.
Our variant analysis also showed that two genetic variants tend to be associated with a lower occurrence of PEP. These two variants in the CPAMD8 and TUBGCP6 genes have not been reported in relation to pancreatitis, and confirmation through a large-scale genome-wide association study is needed.
We also examined whether genetic variants previously known to be associated with pancreatitis were associated with PEP. Among the candidate genetic variants, CASR R990G was identified in the PEP patient group, but there was no significant difference in variant allele frequency in the control group. CASR R990G is a relatively common variant, showing a VAF of 20.6% in the entire 1000 genome database, especially 52% in East Asians, and subjects in this study also showed a similar level of VAF to East Asians in the database [29]. Muddana et al. [35] demonstrated that CASR R990G was more related to chronic pancreatitis than to recurrent acute pancreatitis, especially in subjects who consumed alcohol. In a study of Scillitani et al. [36], CASR R990G showed little effect on serum calcium level. Therefore, the mechanism by which CASR R990G is involved in pancreatitis is presumed to be progression of fibrosis by an unknown mechanism promoted by alcohol rather than by calcium level [18]. Given the mechanism of involvement of the CASR R990G in the occurrence of pancreatitis, the CASR R990G variant is unlikely to be associated with PEP, as in our data.
This study had some limitations. First, the mechanism of how the IRF2BP1 rs60158447 identified in our study is involved in the development of pancreatitis has not been studied. Therefore, it is difficult to conclude that the IRF2BP1 rs60158447 is a definite risk factor for PEP based on the results of the current study, and future studies on the mechanism including the relationship with IRF2 mentioned above are needed. Second, the number of study subjects was small. However, a statistically significant genetic variant associated with PEP was found despite the small number of subjects. Future validation studies with large cohorts are needed to confirm that the newly discovered genetic variant in this study is a real risk factor for PEP, beyond statistical significance. Third, it was difficult to completely adjust the confounding factors. In particular, regarding PEP prevention, prophylactic pancreatic stent insertion was adjusted for through PS matching, but hydration and use of protease inhibitors were not controlled for or analyzed in this study, and thus may have acted as confounding factors. However, we tried to adjust for most clinical factors that could be associated with PEP, and it is unlikely that confounding factors influenced the results. Fourth, to enhance the robustness of the comparison between PEP patients and controls, we excluded patients with asymptomatic hyperamylasemia, which could signify an intermediate state between patients with PEP and normal amylase levels after ERCP. However, this exclusion may have induced selection bias.
In conclusion, this study showed that the IRF2BP1 rs60158447 was significantly associated with PEP in clinically high-risk groups for PEP. Through future verification of study results, this genetic variant can serve as a genetic marker to select high-risk groups for PEP.
KEY MESSAGE
1. This study showed that IRF2BP1 rs60158447 was significantly associated with the occurrence of post-endoscopic retrograde cholangiopancreatography (ERCP) pancreatitis (PEP).
2. This is the first study to suggest a genetic variant associated with PEP.
3. Genetic variants previously known to be associated with the occurrence of pancreatitis did not show a significant association with PEP.
Notes
CRedit authorship contributions
Young Hoon Choi: data curation, formal analysis, methodology, project administration, writing - original draft; Younggyun Lim: formal analysis, methodology, visualization, writing - original draft; Dong Kee Jang: data curation, writing - review & editing; Dong-Won Ahn: data curation, funding acquisition, project administration; Ji Kon Ryu: data curation, writing - review & editing; Woo Hyun Paik: data curation, writing - review & editing; Yong-Tae Kim: data curation, writing - review & editing; Ju Han Kim: formal analysis, methodology, writing - review & editing; Sang Hyub Lee: conceptualization, data curation, writing - review & editing
Conflicts of interest
The authors disclose no conflicts.
Funding
This study was supported by Chung-Ram Research Grant 2018 from the Korean Association of Internal Medicine.