The effect of susceptibility variants, identified in never-smoking female lung cancer cases, on male smokers
Article information
Abstract
Background/Aims
Genome wide and candidate gene association studies have identified polymorphisms associated with the risk of lung cancer in never-smokers. This study was conducted to evaluate the association between 11 polymorphisms identified in female never smokers and the lung cancer risk in male smokers.
Methods
This study included 714 lung cancer patients and 626 healthy controls. The polymorphisms were genotyped using SEQUENOM MassARRAY iPLEX assay or Taq-Man assay.
Results
Two polymorphisms were associated with the risk of lung cancer in male smokers, as in female never smokers. Male smokers carrying the rs4975616 variant allele had a significantly decreased risk of lung cancer (in a codominant model: odds ratio, 0.77; 95% confidence interval, 0.61 to 0.96; p = 0.02). The rs9387478 polymorphism also reduced lung cancer risk in male smokers (in a codominant model: odds ratio, 0.85; 95% confidence interval, 0.73 to 0.997; p = 0.046). In a stratified analysis, the association between these polymorphisms and the risk of lung cancer was predominant in lighter smokers and for cases of adenocarcinoma.
Conclusions
These results suggest that a subset of polymorphisms known to be associated with the risk of lung cancer in female never smokers is also associated with the risk of lung cancer in male smokers.
INTRODUCTION
Lung cancer is the leading cause of cancer-related death. Although the majority of lung cancer cases are related to tobacco smoking, approximately 20% of lung cancer patients have never smoked [1,2]. Radon, asbestos, and indoor air pollution, such as from cooking fumes, are thought to cause lung cancer in never smokers (LCINS) [1].
LCINS exhibits different epidemiological, clinical, and molecular characteristics to those in lung cancer in smokers [2]. The proportion of LCINS is higher in women than in men [3]. Epidermal growth factor receptor (EGFR) mutations and anaplastic lymphoma kinase rearrangements are more commonly found in LCINS, while KRAS proto-oncogene mutations are more frequent in lung cancer in smokers than in never smokers. Genome-wide association studies (GWAS) have identified several genetic variants that have been associated with the susceptibility to LCINS [4-9]. In a previous study, we also found three polymorphisms that may contribute to lung cancer susceptibility in never-smoking Korean females [10].
Although these variants influenced the risk of lung cancer in never-smoking females, the association of these variants with the lung cancer risk in male smokers has not been studied. The pathogenesis of lung cancer in female never smokers and male smokers can be different. We wondered whether these polymorphisms were confined to never smokers and females. So, we investigated the association with lung cancer risk among male smokers, on the other side of female-never smokers. If these polymorphisms also affect the lung cancer risk in male smokers, this suggests that they may be associated with etiology of lung cancer other than smoking. We conducted a case-control study to investigate the association between the variants identified in the GWAS or candidate gene association studies in never-smoking females and the lung cancer risk in male smokers.
METHODS
Study population
This study included 714 lung cancer patients and 626 healthy controls. All of the patients and control subjects were male smokers (current or former). The patients were histologically confirmed to have lung cancer between September 1998 and June 2012 at Kyungpook National University Hospital (KNUH) in Daegu, Korea. Patient blood samples were provided by the National Biobank of Korea–KNUH, which is supported by the Ministry of Health, Welfare, and Family Affairs of Korea. The blood samples for the 626 healthy controls were randomly selected from the pool of healthy volunteers and provided as follows: 436 from the National Biobank of Korea–KNUH, and 190 from the Korean Biobank Project (4851-307, KBP-2011-24) and the Korean Genome and Epidemiology study (4845-302), which is supported by the Korea Centers for Disease Control & Prevention, Republic of Korea. All materials derived from the National Biobank were obtained under protocols that were approved by Institutional Review Boards. This study was approved by the Institutional Review Board of KNUH (Approval No.: KNUHBIO_09-1018), and written informed consent was obtained from all participants.
Polymorphism selection and genotyping
Eight polymorphisms were selected that were identified in never smoker GWAS [4-9]. Three additional polymorphisms: rs10079250 A > G, rs7631358 G > A, and rs13009079 T > C that contributed to lung cancer susceptibility in never-smoking Korean females in our previous study were also included [10]. Among the 11 polymorphisms studied, 10 were genotyped using SEQUENOM’s MassARRAY iPLEX assay (SEQUENOM Inc., San Diego, CA, USA). The rs7631358 G > A polymorphism was genotyped using the Taq-Man assay (Applied Biosystems, Foster City, CA, USA), following the manufacturer’s instructions. The genotyping analysis was performed in a blinded manner with respect to the case/control status. For quality control, approximately 5% of the samples were randomly selected to be genotyped again using a restriction fragment length polymorphism assay by a different investigator, and the results were 100% concordant.
Statistical analysis
Continuous variables, such as age and pack-years, were analyzed using the Student’s t test. A chi-square test was used to compare the differences between the categorical variables, such as the frequencies of the genotypes between the cases and controls. Unconditional logistic regression analysis was used to calculate the odds ratio (OR) and 95% confidence interval (CI), by adjusting for age and pack-years of smoking. In the stratified analysis, individuals were grouped according to age (≤ 61 vs. > 61), pack-years of smoking (≤ 35 vs. > 35), and histology (squamous cell carcinoma vs. adenocarcinoma vs. small cell carcinoma). The Statistical Analysis System for Windows version 9.4 (SAS Institute, Cary, NC, USA), was used for the statistical analyses. A p value of less than 0.05 was considered statistically significant.
RESULTS
The characteristics of the cases and controls are shown in Table 1. All of the participants were current or former smokers and of the male gender. The proportions of current smokers and pack-years of smoking were higher in the case group than in the control group (p = 0.01 and p < 0.001, respectively). These differences were controlled for in the multivariate analyses by adjusting for the pack-years of smoking.
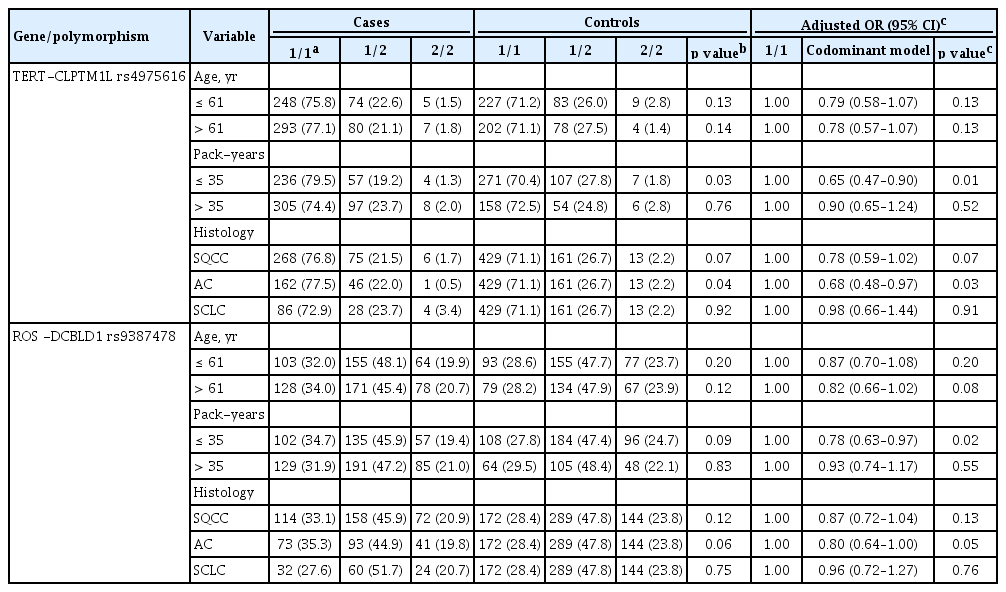
The genotype frequencies of the 11 polymorphisms in the cases and controls are shown in Supplementary Table 1. Among the 11 polymorphisms, three (rs4975616, rs9387478, and rs13009079) showed different genotype frequencies between the cases and controls. After adjusting for age and pack-years of smoking, the rs4975616 A > G polymorphism was significantly associated with the risk of lung cancer. Individuals carrying the rs4975616 variant allele had a significantly decreased risk of lung cancer (in a codominant model: OR, 0.77; 95% CI, 0.61 to 0.96; p = 0.02) (Table 2). The rs9387478 C > A polymorphism was also associated with a decreased lung cancer risk (in a codominant model: OR, 0.85; 95% CI, 0.73 to 0.997; p = 0.046) (Table 2). The rs13009079 T > C polymorphism was related to a risk of lung cancer with only marginal significance (under a dominant model: p = 0.07) (Supplementary Table 2).
In this study, the median value of smoking was 35 pack-years. So we classified cases with less than 35 packyears as lighter smokers and those with more than 35 pack-years as heavier smokers. When stratified by packyears of smoking, the effect of rs4975616 A > G on the lung cancer risk was significant in lighter smokers (OR, 0.65; 95% CI, 0.47 to 0.90; p = 0.01), but not in heavier smokers (OR, 0.90; 95% CI, 0.65 to 1.24, p = 0.52) (Table 3). The effect of rs9387478 C > A on the lung cancer risk was also observed in lighter smokers (p = 0.02). Upon stratification by histological subtype, the association between rs4975616 A > G or rs9387478 C > A and the risk of lung cancer was predominant in adenocarcinoma (p = 0.03 and p = 0.05, respectively) (Table 3). In small cell lung cancer, there was no evidence that either polymorphism was associated with a risk of lung cancer.
DISCUSSION
In this study, we investigated the impact of 11 polymorphisms, which influenced the risk of LCINS, on the risk of lung cancer in male smokers. The rs4975616 A > G and rs9387478 C > A polymorphisms were associated with a decreased lung cancer risk in male smokers.
The rs4975616 polymorphism is located near telomerase reverse transcriptase (TERT) and cleft lip and palate transmembrane protein 1 (CLPTM1L) on chromosome 5p15.33. TERT is one of the major functional subunits of the telomerase enzyme, which plays a critical role in the maintenance of telomere length [11]. CLPTM1L encodes a membrane protein that causes apoptosis when overexpressed in cisplatin-sensitive cells [12]. A number of studies have demonstrated that the TERT-CLPTML1 region of chromosome 5P15.33 is a common susceptibility locus for multiple cancers, including lung cancer [13-17]. A GWAS has reported that rs4975616 A > G is associated with a decreased lung cancer risk in never smokers [6]. In this study, we found that rs4975616 A > G was also associated with decreased lung cancer risk in male smokers. When stratified by pack-years of smoking, the effect of rs4975616 A > G on the risk of lung cancer was significant in lighter smokers, but not in heavier smokers. These results suggest that the rs4975616 variant allele may be associated with a risk of lung cancer, but only in never or lighter smokers.
The rs4975616 A > G polymorphism was not associated with the risk of small cell lung cancer. The effect of rs4975616 A > G on the lung cancer risk was predominant in the patients with adenocarcinoma. A varied effect of rs4975616 A > G, based on the lung cancer histological type, has also been reported in a GWAS of never smokers [6]. In that study, the rs4975616 A > G polymorphism decreased the lung cancer risk in adenocarcinoma, but not in small cell lung cancer [6]. The similar result for the effect of rs4975616 A > G on the lung cancer risk in male smokers, as in non-smokers, suggests that the effect of this variant on the lung cancer risk may not be specific to non-smokers. It was reported that TERTCLPTM1 locus polymorphisms were associated with telomere length [18]. So, there is a possibility that the TERT-CLPTM1L rs4975616 A > G may influence on telomere length and affect lung cancer susceptibility. There may be in linkage disequilibrium between rs4975616 and other functional polymorphisms affecting telomere length. Additional studies are needed to understand the biological mechanism of rs4975616 A > G on lung cancer development.
The rs9387478 polymorphism is located in an interval between ROS proto-oncogene 1, receptor tyrosine kinase (ROS1) and discoidin, CUB and LCCL domain containing 1 (DCBLD1) on chromosome 6p21.2. ROS1 functions as a receptor tyrosine kinase. Increased expression or rearrangement of ROS1 has been reported in nonsmall cell lung cancer [19]. Little is known regarding the functional role of DCBLD1. A GWAS has identified that rs9387478 C > A is associated with a decreased lung cancer risk in never-smoking women in Asia [4]. In that study, rs9387478 was associated with a risk of adenocarcinoma, when analyzed by the histological subtype of lung cancer [4]. In this study, rs9387478 was also associated with a decreased lung cancer risk in male smokers, and the association was significant for adenocarcinoma only. Different EGFR mutation status has been reported, based on the rs9387478 genotypes. In never-smoking female lung cancer cases, the frequency of the rs9387478 AA genotype was significantly higher in the EGFR mutation-negative group than in the EGFR mutation-positive group [20]. Therefore, the varied effects of rs9387478 A > G on the risk of lung cancer, according to the histological subtype, may have been due to the different EGFR mutations. In this study, the association between the rs9387478 genotypes and the EGFR mutation status was not evaluated.
There are several limitations in this study. First, the p value of ROS1-DCBLD1 rs9387478 was less than 0.05 in the codominant model. However, the p value has marginal significance when considering multiple tests. Second, the relationship with survival outcomes is not investigated. For example, the ROS1-DCBLD1 rs9387478 was reported to correlate with overall survival in female nonsmoking patients with lung cancer [20]. However, we could not analyze survival outcomes of male smokers due to lack of survival data. And a lack of functional study may also be a weakness of this study.
Our study has shown that the effects of polymorphisms are not confined to never smokers. Two polymorphisms were associated with lung cancer risk in both never-smoking females and male smokers. Since there are little known about the functional roles of these polymorphisms, the reasons why they are related to lung cancer risk are unclear. Additional studies on the polymorphisms may reveal the etiology of lung cancer other than smoking.
In summary, polymorphisms that are known to be associated with the risk of LCINS were not specific to never smokers. The TERT-CLPTM1L rs4975616 A > G and ROS1-DCBLD1 rs9387478, which were identified in GWAS of never-smoking females, were also associated with the risk of lung cancer in male smokers.
KEY MESSAGE
1. This study was conducted to evaluate the association between 11 polymorphisms identified in female never smokers and the lung cancer risk in male smokers.
2. The telomerase reverse transcriptase (TERT)-cleft lip and palate transmembrane protein 1 (CLPTM1L) rs4975616 and ROS proto-oncogene 1, receptor tyrosine kinase (ROS1)-discoidin, CUB and LCCL domain containing 1 (DCBLD1) rs9387478, which were identified in genome-wide association studies of never-smoking females, were also associated with the risk of lung cancer in male smokers.
Notes
No potential conflict of interest relevant to this article was reported.
Acknowledgements
This study was supported by a grant from the National R&D Program for Cancer Control, Ministry of Health and Welfare, Republic of Korea (grant number: 1720040); the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (No.NRF-2017R1D1A3B03032114).