Interphase Cytogenetics of Lung Tumors Using In Situ Hybridization: Numerical Aberrations
Article information
Abstract
Objectives
Since conventional cytogenetic analysis for bronchogenic carcinogenesis is limited by the difficulty to get enough number of high quality metaphase spreads, the development of new method to overcome above problems is strongly needed. Therefore, the introduction of non-radioactive in situ hybridization (ISH) with pericentromeric chromosome probes gave us the way to investigate the genetic events during carcinogenic process. We applied this method on lung cancer tissue to validate the possibility of this method for general usage and to analyze numerical chromosome aberration status and their clinical correlations.
Methods
A set of satellite DNA probes specific for chromosome 3, 7, 9, 11, and 17 was hybridized directly to paraffin-embedded tissue section of 30 non-small cell lung cancers. Mean chromosome index of each chromosome and frequency of polysomy for each chromosome were calculated.
Results
Mean chromosome indices for chromosome 3, 7, 9, 11, and 17 were 1.10, 1.13, 1.17, 1.12, and 1.17. respectively. Polysomy for a set of chromosomes was detected in all 30 cases except 4 cases which showed hypoploidy only for chromosome 3 or 7 in 2 cases and diploidy only for chromosome 3 or 11 in 2 cases. Among the set of chromosomes, mean chromosome index and polysomy frequency for chromosome 9 & 17 were significantly higher than that for others. Mean chromosome index or polysomy pattern for each chromosome was not much different among cell types or clinical stages.
Conclusions
Our results show that chromosome ISH can be used to screen for numerical chromosome aberrations on paraffin tissue sections and further studies for ISH analysis with different probes on same tumor area or double-target ISH in large scale are needed to confirm above results and to elucidate the specific meanings.
INTRODUCTION
Neoplastic cells are characterized by genomic changes that may be analyzed by cytogenetic, molecular genetic, DNA cytometric or morphometric methods. However, tumor cytogenetic analysis, esp. in solid tumor cytogenetics, is limited by the difficulty to get enough number of high quality metaphase spreads that are representative of the tumorigenic cells in the population or by the genetic alterations which might occur during short-term or long-term culture to obtain mitotic cells1,2). Therefore the introduction of nonradioactive in situ hybridization (ISH) with cloned DNA probes capable of detecting specific repetitive target sequence or whole chromosome library sequence was a major advance in the field of tumor cytogenetics. Furthermore, using this method, it became possible to localize individual chromosome in non-mitotic interphase nuclei3–5).
From comparison between interphase and metaphase studies, it became clear that the number of copies of specific metaphase chromosomes could be evaluated by ISH to interphase nuclei6–10). Also, it became possible to detect several nucleic acid targets simultaneously with different fluorescent colors which are enabled to evaluate structural chromosomal aberration, not only in metaphase but also in interphase nuclei11–13).
Furthermore, the appplication of this method to routine paraffin embedded tumor tissue section might give us the way for the general usage of interphase cytogenetics14–17). In this paper, we investigated the status of numerical chromosomal aberrations in lung cancer tissue using chromosome ISH method and analyzed and correlated to clinical or pathological status.
MATERIALS AND METHODS
Materials
30 cases of primary non-small cell lung cancer (NSCLC) tissue were obtained from routinely buffered formaldehyde fixed paraffin embedded block file and blocks were 3 years old.
Chromosome in situ hybridization
Chromosome in situ hybridization (ISH) was carried out by a previously reported procedure with minor modification17). Briefly, 6 um-thick-tissue sections were mounted on glass slides pretreated with silane or poly-L-lysine, baked overnight at 65°C, and deparaffinized in xylene-ethanol series. These slides were then incubated with 0.4% pepsin in 0.2 N HCl at 37°C for 45–60 minutes after presoaking in a cold pepsin/HCl solution for 15 minutes. The DNA probe was added onto the tissue sections, which were then covered with coverslips, sealed with rubber cement, and denatured together in a wet chamber at 95°C for 3–4 minutes and hybridized overnight at 37°C. Posthybridization washing was done in 50% formamide/1×SSC at 37°C and followed by washing in 0.1×SSC at 37°C.
DNA probes
The repetitive satellite DNA probes detecting sequences of the (peri)centromeric regions of chromosome 3 (D3Z1), 7 (D7Z1), 9 (D9Z5), 11 (D11Z1), and 17 (D17Z1) were purchased in the biotin-labelled form from Oncor (Gaithersberg, MD).
Detection of hybridized signals
Signals were visualized by the immunoperoxidase reaction using the preformed avidin-biotin-peroxidase enzyme complex (Vector, Burlingame, CA) and diaminobenzidine (DAB) as a chromogen. Signal evaluation was performed by counting more than 400 tumor nuclei and more than 200 infiltrating lymphocytes nuclei as an internal control to normalize for tissue truncating effect and variations in ISH efficiencies from slide to slide, within each section. The chromosome index was defined as the ratio of mean number of signals per nucleus of tumor cells to that of infiltrating lymphocytes.
RESULTS
Clinical Informations
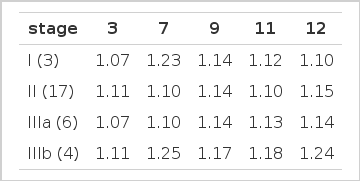
Total 30 cases of surgically resected non-small cell lung cancer specimens were obtained from 17 squamous cell cancer, 9 adenocarcinoma, 3 bronchioloalveolar cell cancer and 1 large cell cancer. 3 cases were stage I, 17 cases were stage II, 6 cases were stage III a, and 4 cases were stage III b (Table 1).
Evaluation of chromosome copy numbers
Mean chromsome number of tumor nuclei for 3, 7, 9, 11, and 17 were 1.75 (1.56–1.85), 1.74 (1.65–2.01), 1.80 (1.59–2.08), 1.75 (1.63–1.86), 1.83 (1.64–2.27), respectively. Mean chromosome indices of tumor nuclei for 3, 7, 9, 11, and 17 were 1.10 (0.96–1.19), 1.13 (0.98–1.28), 1.17 (1.03–1.14), 1.12 (1.00–1.29), 1.17 (0.97–1.47), respectively (Table 2). Distribution of chromosome copy number per nucleus was shown in (Fig. 2). The mean frequency of polysomy (tumor nuclei with 3 or more signals) for 3, 7, 9, 11, and 17 were 6.5%, 8.6%, 13.3%, 7.4%, 12.7%, respectively. But, the meaningful frequency of polysomy (tumor nuclei with polysomy >4%) for 3, 7, 9, 11, and 17 were 76.7%, 70.0%, 100%, 83.3%, 96.7%, respectively (Table 3).
Relationship of chromosome copy number to cell type or clinical stage
Mean chromosome indices for 3, 7, 9, 11, and 17 in squamous cell cancer were 1.10, 1.14, 1.17, 1.12, 1.17; in adenocarcioma were 1.07, 1.10, 1.15, 1.11, 1, 16; in bronchioloalveolar cell cancer were 1.13, 1.13, 1.08, 1.10, 1.22: in large cell cancer were 1.15, 1.21, 1.03, 1.11, 1.17, respectively (Table 4). There was no cell type specific chromosomal change. The mean frequency of polysomy or frequency of polysomy with more than 4% of total tumor nuclei was slight higher in squamous type than in adenocarcinoma (Table 5 & 6) and those differences were not statistically significant.
Mean chromosome indices in each clinical stage were not significantly different for 3, 7, 9, 11 or 17 (Table 7).
DISCUSSION
The cellular DNA content analysis of certain malignancies is regarded as a prognostic parameter. Therefore, the FCM analysis became a rapid and objective screening method for the DNA content of malignant cells18–20). However, no small variations in DNA content can be detected. Cytogenetic analysis of solid tumors is a more precise approach and detects numerical and/or structural chromosomal aberrations2) that might have clinical significance in prognosis and therapy.
In Situ Hybridization is a powerful approach to study numerical chromosome aberrations in cancer cells, as well as cancer cell heterogeneity6,16,17,21,22). As compared to the flow cytometric data that could be obtained, chromosome ISH can give more precise information, and as compared to karyotyping, all cases could be analyzed while also considerably more tumor cells per case can be examined16,17).
Despite the high incidence of non small cell lung cancer (NSCLC), the cytogenetic data available are extremely limited in this neoplasm, compared with those for less frequent hematologic malignancies23). Primary NSCLC specimens often have a low mitotic index, making it difficult to obtain adequate numbers of well banded metaphase cells for detailed cytogenetic analysis. Moreover, the karyotypes can be extremely complex, with many additional chromosomes, complicating efforts to identify consistent changes, i.e. there are several characteristics which make it difficult to assess the extent and significance of numerical chromosomal abnormalities. Those are a wide range of chromosome numbers per cell, lack of a clear modal chromosome numbers, large populations of cells with chromosome numbers in the triploid or tetraploid range, and difficulties in evaluation of structural abnormalities hindered by the large number and complexity of the rearrangements24). Upto now, several groups have reported detailed karyotypic findings in NSCLC from conventional cytogenetic analysis and these studies have revealed multiple numerical and structural aberrations in this neoplasm24–30). For the numerical chromosome aberrations, one of the most common findings is the isochromosome (polysomy) of chromosome 7, 8 or 9. On the contrary, recurrent loss of several chromosomal segments, including 3p, 9p, 11 p, 17p was also reported.
In this paper, we investigated the numerical chromosome aberrations for the chromosome 3, 7, 9, 11, and 17, using chromosome ISH methods on paraffin tissue sections of lung cancer. As for chromosome index, all mean indices are above 1.0 with somehow wide range from 0.96 to 1.47 that means most of tumor cells have polysomy to some extent (Table 2). Indices of chromosome 9 & 17 were higher than that of the others (P <0.05), but the specificity of those changes are needed to be reevaluated with further studies in large scale. As mentioned before, the previous cytogenetic reports showed polysomy of some chromosome including 7, 8, or 9, but for 17 what is one of the deletion site. For the proper explanation of those discrepancies, following assumptions might be used; a) the number of tumor cells being analyzed using ISH method was much higher than that of conventional method (at least 400 tumor cells vs 2–3 metaphase spreads), b) the chromosome copy number might be high, if concurrent deletion of some chromosomal segments excludes centromeric area, because the probe which we used only hybridized to centromeric portion, c) intratumoral heterogeneity.
When we locked into the polysomy frequency of each chromosome, it became more clear that most of tumor cells have high copy numbers of chromosome (polysomy) shown in figure 1 & 2. Even though truncating effect of tissue sectioning, the norm should be expected to have not more than two chromosome copy number for each set of chromosomes. But according to investigators who used same method to detect the numerical chromosomal aberration, even normal lymphocytes might show more than two copy number with range of frequency from 1.2% to 4% of total lymphocytes7,8). So we used the criteria of polysomy when more than 4% of total tumor cells have three or more chromosome copy number. As shown in table 3, mean frequency of polysomy population for each chromosome is above 4%. Mean frequency of chromosome 9 & 17 is much higher than the others (P <0.01), and this tendency is much stronger than in chromosome index.

ISH signals using chromosome 17 centromeric probe on paraffin tissue section of squamous lung cancer. Signal detection with peroxidase/DAB staining and Giemsa counterstain1: most tumor nuclei showed 3 or more signals (polysomy) (original magnification, × 1000)
The chromosomal polysomy for 3, 7, 9, 11, 17 which resulted from above investigations was the first report in lung cancer tissue using in situ hybridization and those results need to be confirmed & reevaluated for the specific meanings through further studies in large scale.
In squamous cell type, mean chromosome indices of each chromosome are similar to that of all cell type and chromosome indices of 9 & 17 are significantly higher than others. But in adenocarcinoma, mean chromosome indices of each chromosome are lower than those of squamous cell type and chromosome indices of 9 & 17 are also siginificantly higher than others. In bronchioloalveolar cell type, mean chromosome index of chromosome 9 is the lowest one (1.08) and that of 17 is much higher (1.22) than others, but statistical analysis could’nt be done because of small number of cases (3 cases).
Relationship of chromosome copy number to different clinical stages was more difficult to analyze statistically. Chromosome index of 17 only was significantly high in advanced stage (III b) and chromosome indices of 9 & 11 showed higher thendency in stage III b but there was no statistical difference.
In conclusion, it can be stated that our investigation for chromosomal aberration analysis with a set of DNA probes shows 1) chromosome ISH can be used to screen for numerical chromosomal aberrations in lung cancer without requiring cell culture system; 2) all chromosomes (3, 7, 9, 11, 17) which we used show some extent of polysomy and polysomies for chromosome 9 or 17 are more frequent & significant; 3) those chromosomal aberations do not show the difference among cell types or clinical stages; 4) further studies for ISH analysis with different probes on same tumor area or double-target ISH in large scale are needed to confirm these results and to elucidate the specific meanings.