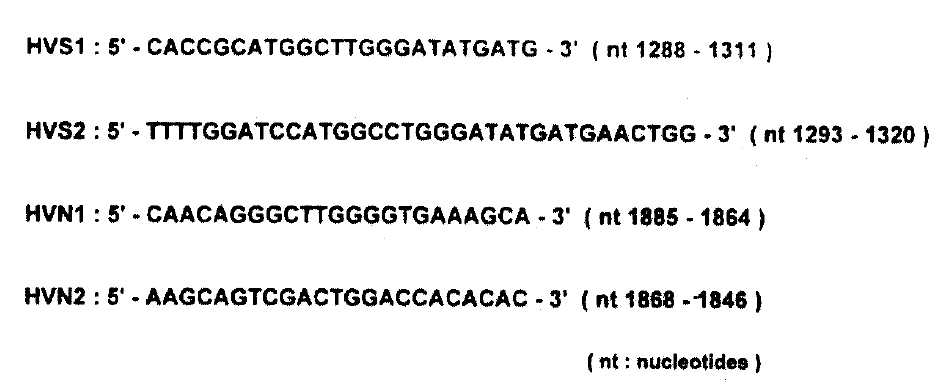Viral Loads and E2/NS1 Region Sequences of Hepatitis C Virus in Hepatocellular Carcinoma and Surrounding Liver
Article information
Abstract
Objectives
Numerous epidemiologic data have documented that chronic infection by hepatitis C virus (HCV) is a major risk factor for the development of hepatocellular carcinoma (HCC). But the molecular mechanism underlying these strong epidemiologic associations between HCV and HCC has not been elucidated. We observed the changes of HCV in HCC to investigate the association of HCV with HCC.
Methods
We used competitive and quantitative polymerase chain reaction and dideoxy-nucleotide chain termination method to compare HCV titers and sequences of the hypervariable region of E2/NS1 region of four isolates from the HCC and surrounding cirrhotic liver tissues in two anti-HCV positive patients.
Results
The copy numbers of HCV-RNA were 1×106 and 4×106/gm wet weight of HCC, and 8×107 and 3.2×108/gm wet weight of cirrhotic liver tissues from patient-1 and -2. The sequence differences between HCV RNA in HCC and in cirrhotic liver were two and five nucleotides in patient-1 and in patient-2 respectively. The amino acid sequences were changed in one and two site in each patient.
Conclusion
These findings may suggest the possible etiological role of HCV in carcinogenesis of HCC, but complete sequence analysis of HCV including multiple isolates in the same patient, should be performed in many cases.
INTRODUCTION
Hepatocellular carcinoma (HCC) is one of the most common malignancy worldwide1) and associated cirrhosis is a major risk factor for this tumor2). Hepatitis C virus (HCV) is a major pathogenic agent of chronic hepatitis which often leads to cirrhosis and HCC3). It is unclear whether the viral infection increases the risk of HCC only through the associated cirrhosis or whether some strains of HCV are directly carcinogenic.
After discovery of HCV and the development of serologic diagnosis4,5), it has been known that about 1% of the general population of the whole world is infected by the virus and many epidemiologic studies demonstrate close association between chronic HCV infection and the development of HCC6). But the molecular mechanisms underlying these strong epidemiologic associations between HCV and HCC have not been elucidated. Recently, it has been reported that HCV genomic RNA existed in HCC and in the surrounding cirrhotic liver tissues and that the sequences obtained from each tissue were different and demonstrated several mutations in same patient. To further investigate the significance of HCV changes in HCC and cirrhotic liver tissues, we quantitated the HCV RNA titers in HCC and the surrounding cirrhotic tissue and sequenced relatively long segment, including hypervariable region (HVR) of the E2/NS1 gene from each sample in two patients.
SUBJECT AND METHODS
1. Patients
Two male, anti-HCV positive patients (56 and 57 years old) with liver cirrhosis and HCC were selected for this study. HCV RNAs were detected in the serum of these two patients. All serologic markers for hepatitis B virus (HBV) were negative. The liver samples were obtained from two patients at surgery for HCC and divided into only cirrhotic liver and only HCC by means of histological examination of frozen sections with special precautions for RNA preparation, and immediately stored at −80°C.
2. Methods
1) Extraction of total RNA from Cirrhotic Tissues and HCC
Total RNAs from cirrhotic tissues and HCC of the patients were extracted by the guanidinium isothiocyanate method7).
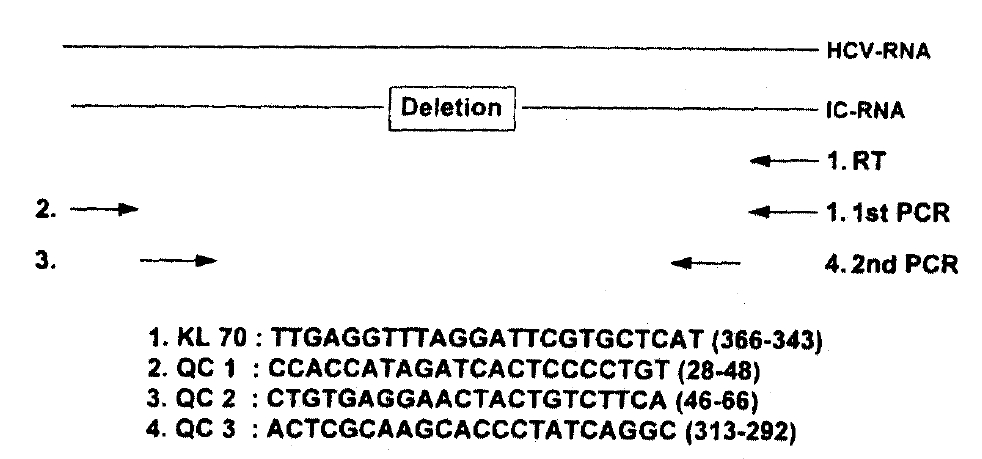
2) Quantitative and Competitive PCR Assay (Fig. 1)
Aliquots of extracted RNA were mixed with twofold serial dilutions of the internal control RNA (10μg/μl) synthesized from 5′-untranslated region of HCV-L125). Subsequently, RNAs were mixed with primer KL70 in a 10μl volume of 1x reverse transcriptase buffer [50mM Tris-HCI (pH8.3), 3mM MgCI2, 75mM KCI, 10mM DTT, 250mM dNTPs] and heated to 70°C for 5 minutes. After quick chilling, 20 units of RNase inhibitor and 100 units of Moloney Murine Leukemia Virus reverse transcriptase (BRL, USA) were added, and the mixture was incubated for 60 minutes at 37°C and then 5 minutes at 100°C. One-tenth of the cDNA was added to the PCR reaction mixture, containing 5pmol of KL70 and QC 1 primers, in a final volume of 20μl [10mM Tris-HCI(pH 9.0), 50mM KCI, 5mM MgCI2, 0.1% Triton X-100, 0.3unit of Taq DNA polymerase, 0.2mM dNTPs]. The mixture was amplified in a DNA thermal cycler (Perkin-Elmer/Cetus, USA, Gene Amp PCR 9600 System) for 40 cycles (950°C for 1 min, 55°C for 1 min, 72°C for 2 min). One-tenth of the first PCR product was added to a second PCR reaction mixture containing nested PCR primers QC 1 and QC 2 and was amplified for 25 cycles as indicated above. Since the internal control RNA contains 79 base deletions, a PCR product derived from the internal control RNA could be separated from that of HCV RNA by gel electrophoresis. By comparing these two PCR products, we were able to quantify HCV RNA.
3) Sequence Analysis of Hypervariable Region (HVR) of the E2/NS1 Gene of HCV
To analyse sequence variation in cirrhotic tissues and HCC, we performed first-strand cDNA synthesis as follows; 500ng of RNA isolated from each liver tissues were mixed with 10μl 1x reverse transcriptase buffer (50mM Tris-HCI (pH8.3), 3mM MgCI2, 75mM KCI, 10mM DTT, 250mM dNTPs) containing 5 pmol random hexamer (5-NNNNNN-3, N is G, A, T or C) and reverse transcriptase reaction was carried out as indicated above. One-tenth of the cDNA was added to the PCR reaction mixture containing 5 pmol of HVS1 and HVS2 primers (Fig. 2) in a final volume of 100μl [10mM Tris-HCI(pH 9.0), 50mM KCI, 5mM MgCI2, 0.1% Triton X-100, 0.3 unit of Taq DNA polymerase, 0.2mM dNTPs], The mixture was amplified for 40 cycles (95°C for 1 min, 55°C for 1 min, 72°C for 2min). The first PCR product (1μl) was transferred to the second-round PCR reaction mixture containing HVN1 and HVN2 primers (Fig. 2) and the same buffer as in first-round PCR, and was amplified for 25 cycles as described above. After the second amplification, PCR product was separated by electrophoresis on a 2% agarose gel and a portion of gel was cut out, corresponding to the position of PCR products with the expected size. DNA was eluted from it and treated with T4 DNA polymerase and T4 polynucleotide kinase (New England Biolabs, U.S.A.). The 5-phosphorylated PCR products were cloned into M13mp18 or-mp19 phage vector and sequenced in both directions by the dideoxynucleotide chain termination method8) by the use of Sequenase version 2.0 (United States Biochemicals). For each other, at least three independent clones were sequenced.
RESULTS
1. Quantitative and Competitive PCR
HCV-RNA titer in HCC and cirrhotic tissues measured by QC-PCR were compared. In patient-1, the copy number of HCV-RNA was 1×106/gm wet weight of HCC and 8×107/gm wet weight of cirrhotic tissue. In patient-2, the copy number of HCV-RNA was 4×106/gm wet weight of HCC and 3.2×108/gm wet weight of cirrhotic tissue. As a whole, the titer of HCV-RNA in HCC was about 10−10 when we compared it with the titer of HCV-RNA in cirrhotic tissue.
2. The Nuceotide and Amino Acid Sequences
The nucleotide and amino acid sequences of hypervariable region in HCC were compared with that of HVR in cirrhosis of patients-1 and -2. The region sequenced here corresponds to nucleotides -1289 to -1798 in patient-1 and -1363 to -1589 in patient-2. In patient-1, two nucleotide and one amino acid sequence in HCC were different from that in cirrhosis. In patient-2, five nucleotides and two amino acids sequences were changed in HCC (Fig. 3 and 4). The sequences obtained from two independent amplifications of the same sample were identical.
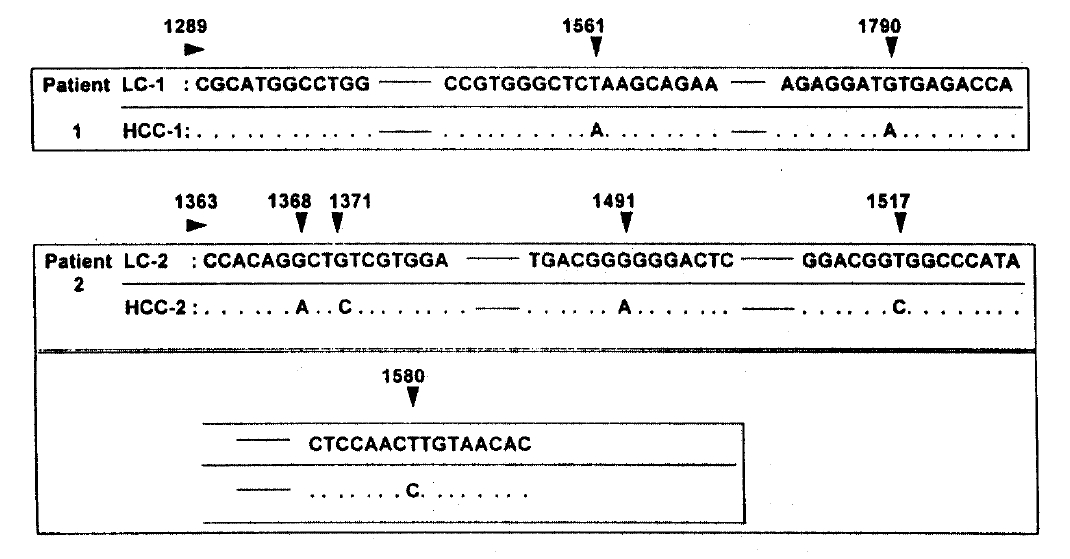
Nucleotides sequences of the hypervariable region in liver cirrhosis (LC) and Hepatocellular carcinoma (HCC) tissues from two patients. Solid line denotes the omitted, identical sequences in LC and HCC. Dotted line denotes the same sequences as the above.
DISCUSSION
HCC is the eighth most frequently reported malignancy worldwide1) despite the striking geographical and racial variations in its prevalence. The association between HBV and HCC has been clearly documented in HBsAg-positive patients2,9,10). Chronic HCV infection has now emerged as another potentially important factor in liver cancer6).
The prevalence of HCV infection varies in distinct geographical areas and it ranges from 0.1%11) – 5.2%12). Current estimates suggest that up to 300 million people worldwide have been, or are, infected with HCV and HCV causes chronic infection in 50–80% of those infected and may ultimately lead to severe liver disease, cirrhosis and HCC13). But the molecular bases for the higher rate of HCC development in patients with chronic HCV infection has not been elucidated.
To confirm the fact that a specific virus induces a human malignancy, the virus should be detected and confirmed in the cancer tissue. We detected the HCV genomic RNA in tumorous and nontumorous tissues from two anti-HCV positive patients. The possibility of contamination of the tumor by serum HCV particles or nontumorous cells was definitely eliminated by the comparison of HCV RNA titers and sequences from each sample after detection of HCV RNA. From each sample, the copy numbers and sequences obtained were different and reproducible in three independent amplifications. The copy numbers of HCV RNA in HCC tissues from two patients were much lower than that of HCV RNA in the surrounding cirrhotic liver tissues (about 10−10) but we did not investigate whether HCV genomes are present at low copy number per cell or only a limited number of tumor cells contained the viral RNA. The lower number of copies of viral genome in HCC may suggest the possible role of the so-called “hit and run mechanism” as it has been widely observed and suggested in other tumors14–18). The changes of viral structures and replication capacity in tumor i.g. newly developed and different environment from surrounding tissue may be suggested, but it is not certain whether these changes of virus occurred in an already developed tumor or occurred before development of the tumor.
The HVR at E2/NS1 gene of HCV is highly divergent between different variants of HCV19–21) and show the highest rate of sequence change with time in persistently infected individuals22,23). As these proteins are likely to lie on the outside of the virus, they would be the principal targets of the humoral immune response to HCV. It has been suggested that these proteins are involved in the virus-cellular receptor interactions which are, therefore, the most susceptible to neutralization and under the greatest pressure to mutate24,25). Therefore, we selected a portion of this region for sequence analysis and comparison of the HCV genomes isolated simultaneously from HCC tissue and the surrounding cirrhotic liver in the same patients. We analyzed relatively long sequences (about 600 nucleotides) including HVR of four isolates from two patients.
The sequences obtained from HCC and cirrhotic tissue in the same patients demonstrated some changes of nucleotides and aminoacid sequences. These differences may be the cause or result of HCC development i.g. and the viral changes may be related with cancer development or contrarily the virus changed in the new environment after cancer development. Because we did not isolate the multiple genome from each specimen, we could not exclude the possibility of the coexistence of quasispecies of the virus in the same patient. But the complete sequence analysis of the HCV including multiple isolates in the same patient should be performed in many cases because the minimal change (s) of the virus could induce the malignant transformation.