Gut microbiota dysbiosis and its impact on asthma and other lung diseases: potential therapeutic approaches
Article information
Abstract
The emerging field of gut-lung axis research has revealed a complex interplay between the gut microbiota and respiratory health, particularly in asthma. This review comprehensively explored the intricate relationship between these two systems, focusing on their influence on immune responses, inflammation, and the pathogenesis of respiratory diseases. Recent studies have demonstrated that gut microbiota dysbiosis can contribute to asthma onset and exacerbation, prompting investigations into therapeutic strategies to correct this imbalance. Probiotics and prebiotics, known for their ability to modulate gut microbial compositions, were discussed as potential interventions to restore immune homeostasis. The impact of antibiotics and metabolites, including short-chain fatty acids produced by the gut microbiota, on immune regulation was examined. Fecal microbiota transplantation has shown promise in various diseases, but its role in respiratory disorders is not established. Innovative approaches, including mucus transplants, inhaled probiotics, and microencapsulation strategies, have been proposed as novel therapeutic avenues. Despite challenges, including the sophisticated adaptability of microbial communities and the need for mechanistic clarity, the potential for microbiota-based interventions is considerable. Collaboration between researchers, clinicians, and other experts is essential to unravel the complexities of the gut-lung axis, paving a way for innovative strategies that could transform the management of respiratory diseases.
INTRODUCTION
The human body hosts a diverse microbiota that forms a symbiotic relationship with its host, creating a complex microecosystem. Maintaining a balanced microbiota is crucial as imbalances can lead to aberrant immune responses and inflammation. Specifically, the gut microbiota refers to a diverse community of microorganisms residing in the gastrointestinal tract, including bacteria, fungi, protozoa, archaea, and yeasts [1].
The gut, initially sterile during intrauterine development, undergoes colonization immediately after birth, with significant fluctuations in bacterial numbers and species occurring during early life [2]. In adults, the gut contains approximately 1014 bacteria, two-thirds of which are specific to an individual. In normal subjects, the gut harbors approximately 500 different species of microorganisms, collectively weighing approximately 1.5 kg [3,4]. Dominant genera of the healthy intestinal microbiome include Clostridium, Faecalibacterium, Ruminococcus, Roseburia, Eubacterium, Bifdobacterium, Prevotella, and Bacteroides [5].
Given its critical role in maintaining overall health, the gut microbiota has garnered significant attention as a potential therapeutic target. This heightened interest stems from two main factors. First, technological advancements have enabled comprehensive detection and analysis of intestinal bacterial flora, providing detailed insights into its composition and pathological variations. Second, the composition and diversity of gut microbiota can be influenced by various environmental and host immunological factors [6], making external manipulation relatively feasible through oral intake of probiotics, antibiotics, and other agents.
The significant influence of the gut microbiota extends beyond gastrointestinal health, impacting several aspects of human health, ranging from appetite and energy metabolism to immunity [7]. For instance, the gut microbiota plays a crucial role in fermenting non-digestible substrates, including dietary fibers and endogenous intestinal mucus, which, in turn, support the growth of specific microbes capable of producing short-chain fatty acids (SCFAs). An increasing body of evidence also suggests a close link between the gut microbiota and lung health [8]. An imbalance between the gut and lung microbiomes could potentially contribute to altered immune function and the development of chronic airway diseases. However, whether this imbalance is the cause or effect of the disease remains uncertain.
This review aimed to elucidate the significant connection between gut microbiota dysbiosis and lung diseases, focusing in particular on asthma. Furthermore, we explored potential therapeutic approaches for modulating the gut microbiota to mitigate lung inflammation. Through a comprehensive review of this emerging field, we report promising avenues for managing lung diseases through interventions targeting gut microbiota.
GUT MICROBIOME AND PULMONARY DISORDERS
Recent studies have reported an association between gut microbiota changes and lung inflammation [8]. Distant inflammatory responses resulting from a “leaky gut” and microbiome changes may contribute to chronic airway diseases, and vice versa [9]. However, there is a lack of data regarding how lung dysbiosis affects gut dysbiosis in the bidirectional interaction of the lung-gut axis, particularly compared to the reverse interaction, i.e., the effect of gut dysbiosis on lung dysbiosis. An experiment evaluating the impact of type-2 lung inflammation on the lung and gut microbiomes in mice overexpressing lung-specific interleukin (IL)-13 demonstrated that microenvironmental changes in the lung with profound type-2 inflammation affect not only the lung but also the gut microbiome [10]. Using animal models, researchers have investigated specific gut microbial compositions and their interactions with the host, contributing to our understanding of the processes underlying gut-lung communication [7]. However, there is a need for further large-scale and longitudinal integrative studies on adult asthma, including all members of the gut microbiome, such as the bacteriome, fungiome, and virome, along with their products.
Asthma
Asthma, a prevalent chronic airway disease affecting over 300 million individuals worldwide, holds significant medical importance [11]. Recent research has provided evidence of the involvement of commensal bacteria in asthma through studies conducted on germ-free mice. These studies revealed that germ-free mice exhibited an exaggerated susceptibility to allergic responses, indicating a pivotal role of commensal gut bacteria in regulating immune responses associated with asthma [12,13]. Over the past 15 years, advancements in high-throughput sequencing have provided valuable insights into the significant impact of the gut microbiome on asthma, particularly during the critical period spanning the first 2 years of life [8]. Disruptions in the normal gut microbiota composition during this critical period, attributed to external factors, such as antibiotic exposure, Cesarean-section delivery, shorter breastfeeding duration, and modern residential environments, have been associated with an increased risk of developing asthma and allergic diseases [14,15]. Compositional alterations in gut microbiota during this period, characterized by reduced abundance of the genera Akkermansia, Bifidobacterium, and Faecalibacterium, are potential predictors of asthma and allergic diseases [16]. A Canadian neonatal longitudinal cohort study identified four taxa (Faecalibacterium, Lachnospira, Veillonella, and Rothia, together abbreviated as FLVR) exhibiting a protective effect against asthma during the first 100 days of life [17]. Germ-free mice supplemented with FLVR taxa were also found to be protected from allergic airway inflammation, exhibiting increased fecal concentrations of acetate, a SCFA known to protect against asthma. However, there have been fewer studies investigating the gut-lung axis in adult asthma compared to childhood asthma. A pilot study examining the adult asthma phenotype provided compelling evidence of a robust correlation between forced expiratory volume in one second and phylum-level differences in gut bacterial composition. Notably, alterations in the relative abundances of Bacteroidetes and Firmicutes phyla, such as a lower Bacteroidetes to Firmicutes ratio, were observed, suggesting a possible association between gut microbial composition and lung function in asthma patients [18].
Several studies have highlighted the significance of specific gut microbes, particularly beneficial probiotics, including the Lactobacillus and Bifidobacterium genera, in mitigating allergic asthma, both in animal and human studies [19-21]. Oral exposure of adult mice to house dust has demonstrated that Lactobacillus enrichment in the intestinal microbiome, resulting in enhanced airway immune defense against allergens and viral infections, was based on the negative regulation of Th2 inflammation by the abundance of Lactobacillus johnsonii [19]. In human infant studies, colonization by Lactobacilli and Bifidobacteria was found to correlate inversely with the risk of allergy [20]. In a murine model of chronic asthma, oral supplementation with specific microbes triggered an anti-inflammatory response. Bifidobacterium breve suppressed airway inflammation by inhibiting both neutrophil and eosinophil lung infiltration [22,23]. Clostridium species, dominant in the distal small intestine and colon, have been shown to increase the proportion of regulatory T cells (Tregs) in mice [24-27]. For instance, oral administration of Clostridium leptum in adult mice has shown a negative correlation with asthma [28]. Oral feeding with C. leptum for 2 weeks increased the percentage and total number of Tregs in the spleens and mediastinal lymph nodes, and enhanced IL-10 and transforming growth factor-β1 production in the lungs. A recent study demonstrated a negative association between asthma severity and gut Akkermansia muciniphila in both human and mouse models [29]. These findings suggest that specific gut microbiota species may regulate asthma by modulating airway inflammation.
A proposed mechanism linking gut microbiota to the reduction in allergic airway diseases involves the secretion and disturbance of metabolites that suppress lung inflammation [30,31]. SCFAs, such as butyrate and propionate, are recognized for their pivotal role in the gut-lung axis [32]. The gut microbiota exerts a unique influence on the host’s bile acid metabolism by facilitating processes, such as deconjugation and dihydroxylation, leading to primary and secondary bile acid production. Both primary and secondary bile acids activate the farnesoid X receptor and G protein-coupled bile acid receptor-1 (GPBAR1, also known as TGR5). Secondary bile acids, in particular, significantly impact bacterial translocation and immune regulation through TGR5 [33]. Consequently, gut microbiome dysbiosis can significantly alter the bile acid profiles, thereby influencing asthma pathogenesis through the regulation of both innate and adaptive immunity, particularly in obesity-induced asthma [34].
In vitro and mouse studies have suggested that bile acids could potentially reduce eosinophilic airway inflammation, possibly by promoting Th1 cytokine generation by dendritic cells via the nuclear farnesoid X receptor of these cells [35]. Additionally, an inhibitory effect on the inflammatory unfolded protein response of bronchial epithelial cells has been suggested [36]. A human study reported a significant increase in the levels of taurocholate, a primary bile acid, in children with asthma when compared to healthy individuals [37]. This finding was also replicated among adult individuals with T2-high asthma, with higher levels observed for both taurocholate and glycodeoxycholate [38]. Another possible mechanism that needs further investigation is the gut secretion of soluble IgA. Secretory IgA (SIgA) serves as the first line of defense in protecting the intestinal epithelium from enteric toxins and pathogenic microorganisms [39]. SIgA is known to be induced by either food antigens or intestinal microbiota [40]. Meanwhile, IgA also regulates the composition and metabolic function of gut microbiota [41]. Based on the presence of virulent strains of Streptococcus pneumoniae or Haemophilus influenzae, these interactions may control the local and systemic SIgA levels by producing IgA-specific proteases, thereby influencing respiratory tract infections [42]. Reduced SIgA levels have been observed in asthma and chronic obstructive pulmonary disease (COPD), possibly affecting disease progression [43].
Current research efforts are focused on the association between gut microbiota and distinct subsets of asthma endotypes. For instance, symptomatic eosinophilic adult asthma patients have exhibited altered gut microbiome compositions [44]. Two recent studies have highlighted the correlation between a dysbiotic gut microbiome and asthma phenotypes. Eosinophilic asthma patients showed reduced Clostridia, Lachnospiraceae, and Oscillospiraceae populations, along with an increased Prevotella population in the gut, suggesting an association with bile acid and lipopolysaccharide metabolism [44]. A follow-up conducted at approximately 37.9 days after stepping up the asthma treatment did not reveal any significant difference in gut bacteria diversity and composition in asthma patients. In another study involving 125 adult asthma patients, gut microbiome data were subjected to clustering analysis, resulting in the identification of three distinct enterotypes [45]. The genus Prevotella was found to predominate in the T2-high asthma cluster, characterized by significantly elevated serum periostin, whereas the genera Clostridium and Romboutsia were prevalent in the T2-low asthma cluster, characterized by a high proportion of serum interferon (IFN)-γ.
COPD
COPD, a progressive inflammatory lung disease characterized by airflow obstruction, ranks as the third leading cause of mortality [46]. Emerging evidence suggests that the gut-lung axis may play a crucial role in COPD pathogenesis [47].
In a study comparing the gut microbiome between COPD patients and healthy controls [48], 16S rRNA gene sequencing revealed significant differences in microbial composition. COPD patients exhibited an increased abundance of Streptococcus, Rothia, Romboutsia, and Intestinibacter genera from the family Peptostreptococcaceae and Escherichia in their guts. Conversely, Bacteroides, Roseburia, and Lachnospira genera from the family Lachnospiraceae, as well as several unnamed Ruminococcaceae genera, were decreased in COPD patients. Some specific species, such as Streptococcus sp000187445, Streptococcus vestibularis, and various members of the family Lachnospiraceae, were also found to correlate with reduced lung function.
Since smoking is a major risk factor for COPD development, it is essential to consider its impact on the gut microbiome composition. A cohort study reported gut microbiota changes in male smokers [49], with current smokers showing a higher proportion of Bacteroidetes and lower proportions of Firmicutes and Proteobacteria in their gut microbiota compared to those who had never smoked or had quit smoking. Meanwhile, gut microbiota compositions of individuals who had never smoked and former smokers did not show significant differences. These findings suggest that smoking cessation could potentially facilitate the restoration of gut microbiota to a state resembling that of non-smokers.
Various animal models have been used to investigate the role of the microbiome in COPD [50]. Among these, the cigarette smoking (CS) model uniquely sheds light on the potential influence of the gut microbiome on COPD progression. Interestingly, fecal microbiota transplantation (FMT) from COPD patients (GOLD stage III–IV), exhibiting significantly reduced SCFA levels, into CS murine models aggravated the lung condition [51]. Remarkably, oral administration of antibiotics has demonstrated the capacity to mitigate CS-induced COPD pathogenesis. This intervention yielded a significant reduction in CS-related COPD progression. Within this context, Parabacteroides goldsteinii has emerged as a crucial protective species against COPD [52].
A recent Finnish longitudinal cohort study investigated associations between the occurrence of chronic airway disease and the gut microbiome [53]. The study revealed associations between COPD onset and an increase in Faecalicatena, Oscillibacter, Lawsonibacter, Flavonifractor, and Streptomyces, along with a decrease in Lachnospira, ER4, KLE1615, Eubacterium, and Coprococcus. Notably, the gut microbiome exhibited a higher predictive value for incident COPD compared to asthma (area under the curve: 0.780 and 0.593, respectively). Additionally, studies of fecal samples and monitoring for chronic airway diseases in adults have demonstrated that gut dysbiosis precedes respiratory symptoms, indicating its potential role in COPD etiopathogenesis.
Interstitial lung disease
Interstitial lung disease (ILD) refers to a broad spectrum of airway diseases characterized by lung fibrosis and inflammation [54]. Its cause remains elusive, but recent studies have suggested the potential role of the gut and lung microbiome in fibrotic lung disease. The lung microbiome in ILD differs significantly from that of healthy individuals [55,56]. Idiopathic pulmonary fibrosis (IPF) patients exhibit elevated lung bacterial loads, including the genera of Staphylococcus, Streptococcus, Campylobacter, and Stenotrophomonas, which may contribute to the exacerbation of IPF [55,56]. Currently, there are no relevant data on the gut microbiome of ILD patients.
In the bleomycin-induced pulmonary fibrosis model, germfree mice were protected against lung fibrosis morbidity, with increased gut microbial diversity associated with better fibrosis outcomes [57]. Additionally, the relative abundance of some intestinal probiotics, including Catenibacterium and Lactobacillus (L. johnsonii and L. gasseri), were significantly reduced, while the relative abundance of Verrucomicrobiales and Enterobacteriales were significantly increased in the bleomycin mice model.
TARGETING THE GUT MICROBIOTA
The gut-lung axis has emerged as an important area of research in airway diseases, prompting investigations into various therapeutic approaches to target the gut microbiota and its potential influence on lung disorders. It is no longer sufficient to merely explore associations between the gut and/or lung microbiome and disease outcomes; instead, researchers are now focusing on understanding the complex ecological interactions between the microbiota and the host that influence the immune response.
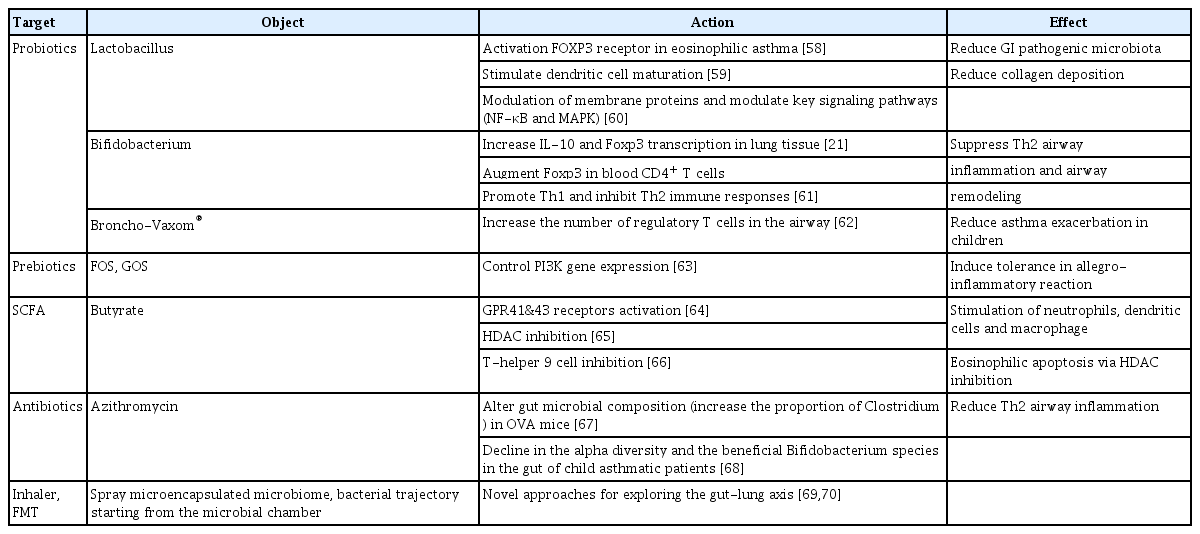
Recent studies have recognized the potential of correcting gut-lung dysbiosis in treating lung disorders. Reviews of preclinical and clinical studies of various interventions, including probiotics/prebiotics, FMT, antibiotics, and metabolites, have reported promising results in modulating the gut microbiota to improve lung health (Table 1) [21,58-70]. However, the mechanisms by which these potential treatment options overcome adaptation and restore the inherent variability in the gut-lung axis are complex and remain poorly understood. The human microbiome is highly dynamic, with its composition changing in response to various factors, including diet, lifestyle, and environmental exposures. Moreover, the gut-lung axis involves intricate interactions between the gut microbiota, immune system, and lung, making it a challenging area to unravel.
Probiotics and prebiotics
Probiotics (i.e., live bacteria administered orally to facilitate intestinal colonization) and prebiotics (i.e., non-viable substrates selectively utilized by host microorganisms to confer health benefits) have been widely used for decades, particularly in infants, to prevent allergic diseases [71]. Probiotic administration may alter the composition of intestinal microbiota and improve microbial balance in the gut [72]. Well-known probiotic strains, including genera Lactobacillus and Bifidobacterium spp., ferment oligosaccharides in the colon, leading to SCFA production. Other potential probiotic species from Clostridium clusters 4 and 14, including C. leptum, Ruminococcus bromii, Faecalibacterium prausnitzii, Coccoides, Eubacterium rectale, Roseburia spp., and Butyrivibrio fibrisolvens, have also shown beneficial effects [73]. These probiotics increased the production of type-1 cytokines, including tumor necrosis factor (TNF)-α and IFN-γ, and decreased the production of type-2 cytokines, including IL-4, in in vivo models [74,75]. Additionally, recent studies have suggested a potential role for extracellular vesicles (EVs) derived from Lactobacillus and Lactococcus. For instance, intranasal administration of EVs isolated from Lactococcus lactis in an experimental model of asthma resulted in reduced airway hyper-responsiveness and type-2 inflammation, indicating that EVs may mediate immunomodulatory effects in asthma [76]. These immunomodulatory effects are particularly relevant in allergic diseases, where an imbalance between type-1 and type-2 immune responses is common. However, it is important to note that probiotics may persist in the adult gut for only a few days, necessitating an individualized approach to enhance intestinal adhesion, particularly in adults [77]. Researchers have explored different strategies to improve probiotic colonization in the gut, to maximize their beneficial effects. A study in mice reported that gut inoculation with L. johnsonii significantly reduced Th2 inflammatory responses in the lungs [19].
Prebiotics include non-digestible soluble fibers, such as fructo-oligosaccharides, galacto-oligosaccharides, and polysaccharides [69]. These dietary fermentable fibers can influence allergic lung inflammation in mice by altering the gut microbiota, leading to increased circulating SCFA levels [65]. Currently, the combination of probiotics and prebiotics, termed “synbiotics”, is used to enhance their favorable effects therapeutically and harness the therapeutic potential of the gut microbiota [78].
Antibiotics
Antibiotics are recognized for their ability to induce compositional changes in the intestinal microbiota. Soluble TNF receptors (TNFR1 and TNFR2) were found to be significantly increased in the sputum of T2-low asthma patients [79]. However, long-term azithromycin treatment significantly reduces sputum TNF and TNFR2 concentrations in non-eosinophilic T2-low asthma patients, compared to healthy controls [80]. Azithromycin is considered a potential treatment for airway inflammatory diseases. A recent study demonstrated that azithromycin modified the gut microbial composition and mitigated allergic airway inflammation induced by FMT in a mouse model [67].
The impact of antibiotic use on childhood asthma is a topic of debate. Studies of children aged 2–7 years have demonstrated that macrolide use leads to increased populations of the phyla Bacteroidetes and Proteobacteria, while decreasing Actinobacteria populations, thereby increasing the risk of asthma and overweight [81]. Therefore, the effects of antibiotic administration on the microbiome, along with the optimal dosage and duration of use, remain controversial, necessitating additional longitudinal studies.
Metabolites
The human gut microbiome has several functions, including catalyzing the metabolism of complex carbohydrates and producing SCFAs [82,83]. Additionally, it contributes to tryptophan metabolism [84] and the production of anti-inflammatory lipids [85], which are crucial for maintaining gastrointestinal health. These metabolic products constitute an essential energy source for gastrointestinal epithelial cells.
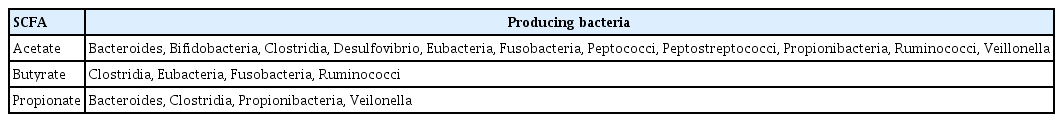
Among the significant metabolites produced by the gut microbiota, SCFAs generated from microbial fermentation of dietary fibers, including acetate, butyrate, and propionate, serve as an energy source for colonocytes [86]. Acetate and propionate are present in both small and large intestines, while butyrate is primarily found in the colon. Changes in SCFA levels can indicate host dysbiosis or bacterial infection, making them valuable indicators of bacterial activity influenced by factors such as dysbiosis, diet, lifestyle, and age [87]. The SCFA type produced depends on the bacteria involved in fermentation (Table 2) [73], and the pathways for production of these substrates differ among species.
Despite evidence from murine studies that oral SCFA administration can alleviate allergic inflammation [32,88], its effect on human allergic diseases remains controversial. The PASTURE study reported that children fed a diet composed of yogurt, fish, vegetables, and fruits exhibited increased fecal butyrate levels, along with a decreased risk of sensitization to food and inhalant allergens [32]. Additionally, females with higher gestational fecal SCFA levels were less likely to have offspring with atopic asthma [89]. Histamine, another important gut metabolite, can regulate NLRP6 inflammasome and intestinal IL-18 secretion, thereby influencing the expression of colonic anti-microbial peptides [90]. In another study, supplementation with Lactobacillus reuteri, a bacterium producing histidine decarboxylase, resulted in the suppression of colonic inflammation by converting L-histidine to histamine in the gut [91]. However, recent studies have indicated that SCFAs induce the release of TNF-α and IL-6 in lung mesenchymal cells, fibroblasts, and smooth muscle cells, suggesting proinflammatory effects rather than anti-inflammatory effects. This underscores the need for further human studies [92].
FMT
The clinical efficacy of FMT, a technique involving the transfer of fecal matter from a healthy donor to a recipient, has been demonstrated in randomized controlled trials for various conditions, including Clostridium difficile infections [93], inflammatory bowel disease [94], obesity [95], type-1 diabetes mellitus [26], and autism spectrum disorder [96]. While FMT has not been established as a therapeutic option for airway diseases due to its high cost and technical complexity, recent research has highlighted its potential impact on lung health. Studies have reported that FMT with feces rich in Bacteroides fragilis from adult asthma patients, when transplanted into gnotobiotic mice, induces a Th17 response in murine airways [97]. In animal models of other respiratory diseases, FMT has been reported to alter the immune cell profile, particularly in cases of broad-spectrum antibiotic treatment or germ-free conditions [98,99]. FMT through selective microbiota transplantation using innovative strategies and ensuring compatibility with the recipient’s gut microbiome holds promise for improving the management of chronic airway diseases.
Future therapeutic prospects: inhalational approach
The unique microbiome of lungs has been subjected to extensive scrutiny, revealing its intricate relationships with both pulmonary equilibrium and lung diseases. A previous study analyzing nasal blow samples from asthmatic children suggested that the Moraxella cluster poses the highest risk [100]. Regarding phylum-level abundance, dysbiosis characterized by increased Proteobacteria or decreased Bacteroidetes levels, has been reported in asthmatic airways compared with healthy controls [101]. In a study involving asthmatic patients with varying severities, the genus Selenomonas was significantly reduced in asthma, correlating with its severity. Furthermore, intranasal pre-treatment with Selenomonas effectively reduced Th2 airway inflammation and airway hyper-responsiveness in an in vivo murine asthma model. This suggests the potential for inhalational therapeutic approaches in modulating lung dysbiosis and related immune responses [102].
Within the context of chronic rhinosinusitis, topical prebiotic administration as a nasal spray was well-tolerated, although it did not significantly impact symptom severity and the microbiological flora [103]. Another study focusing on topical Roseomonas mucosa found a significant reduction in disease severity, the need for topical steroids, and the burden of Staphylococcus aureus colonization in pediatric atopic dermatitis patients [104]. Although these studies investigated other allergic diseases, their findings contribute to establishing a potential causal and mechanistic foundation for the encouraging initial results observed with topical microbiome transplantation in the context of allergic airway disease. Theoretically, coating or microencapsulating certain strains and using them in an inhaled form appears to be appropriate in terms of viability and stability. Similarly, the concept of “mucus transplants”, analogous to FMT, has been proposed. These emerging concepts highlight the importance of continued research to unravel the complexities of leveraging the gut microbiome as a therapeutic strategy in various respiratory conditions.
PERSPECTIVES
The gut microbiome plays a crucial role in maintaining immune homeostasis throughout the body. Disruptions in its equilibrium can impact lung immune function, potentially leading to respiratory disorders, including asthma. A substantial body of evidence suggests a bidirectional regulatory relationship between intestinal commensals and lung inflammation. Alterations in the intestinal microbial milieu and their metabolites can influence the development and progression of respiratory disorders via immunological pathways. By consolidating recent scientific advancements, this review aimed to clarify the potential link between the gut microbiome and asthma, as well as the gut microbiome’s intermediary role in this dynamic, and assess the feasibility of novel therapeutic approaches for modulating gut dysbiosis.
However, understanding the complexity of the microbial landscape is crucial before implementing therapeutic strategies. Studies have reported that approximately two-thirds of the human gut microbiota exhibit individual variations, influenced by factors, such as diet, host genotype, lifestyle, and the use of antibiotics and other medications [105,106]. These variations contribute to the inconsistent gut microbial patterns observed in patients with lung diseases. Additionally, given the heterogeneity of pulmonary diseases, it is essential to consider the interactions of microbiome alterations within these patients [107]. Future research efforts should consider various clinical factors complicating the interpretation of microbial patterns. A comprehensive understanding is essential for developing tailored and effective therapeutic strategies targeting the microbiome. Further mechanistic studies using advanced molecular approaches and longitudinal human studies may enhance our understanding of how changes in gut immune responses induce alterations in respiratory immunity.
Acknowledgements
The figure was created through BioRender.com.
Notes
CRedit authorship contributions
Young-Chan Kim: writing - original draft, visualization; Kyoung-Hee Sohn: writing - original draft; Hye-Ryun Kang: conceptualization, writing - review & editing, supervision, project administration, funding acquisition
Conflicts of interest
The authors disclose no conflicts.
Funding
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (No. RS-2023-00217157).