Infection of human intestinal epithelial cells by invasive bacteria activates NF-κB and increases ICAM-1 expression through NOD1
Article information
Abstract
Background/Aims
Nucleotide-binding oligomerization domain 1 (NOD1) is required for primary intestinal epithelial cells (IECs) to respond to natural mucopeptides secreted by gram-negative bacteria. Infection of human IECs with invasive bacteria up-regulates intercellular adhesion molecule-1 (ICAM-1) expression. However, the role of NOD family members in host defense has been largely unknown. The aim of this study was to determine whether there is a functional role for NOD1 in the up-regulation of ICAM-1 expression in invasive bacteria-infected IECs.
Methods
ICAM-1 mRNA expression was compared between controls, Caco-2 or HT29 cells transfected with an empty vector, and IECs stably transfected with a dominant-negative (DN) NOD1. Expression was compared using qualitative reverse transcription polymerase chain reaction (RT-PCR), real-time RT-PCR, and flow cytometry after infection with enteroinvasive Escherichia coli O29:NM or Shigella flexneri. Nuclear factor kB (NF-κB) activation was determined by electrophoretic mobility shift assays.
Results
DN NOD1 significantly inhibited the up-regulation of ICAM-1 expression in response to an enteroinvasive bacterial infection. The Caco-2 cells transfected with DN NOD1 manifested marked inhibition of NF-kB activation in response to E. coli O29:NM infection.
Conclusions
Signaling through NOD1 may play an essential role in neutrophil trafficking following infection with enteroinvasive bacteria.
INTRODUCTION
Bacterial pathogens that infect the gastrointestinal tract, including Salmonella, invasive Escherichia coli, Listeria, Shigella, and Yersinia spp., are one of the major causes of morbidity and mortality worldwide [1]. The inflammatory program of epithelial cells in response to invasion by bacteria includes the up-regulated expression and production of proinflammatory and chemoattractant cytokines such as tumor necrosis factor α (TNF-α), interleukin 8 (IL-8), and granulocyte macrophage colony stimulating factor (GM-CSF) [2]. The nuclear factor kB (NF-κB) transcription factor is a central regulator of these intestinal epithelial cells (IECs) innate immune responses that are induced by enteroinvasive bacterial infection [3].
Because of many recent studies, it is well known that innate immune responses can be initiated by a system of structurally related proteins that act as pattern recognition receptors (PRRs), termed Toll-like receptors (TLRs), for pathogen-associated molecular patterns (PAMPs) [4]. In addition to TLRs, epithelial cells have intracellular sensors that can response to microorganisms or their by-products. These sensors include some members of the NLR (nucleotide-binding domain and leucine rich repeat containing) family, which includes nucleotide-binding oligomerization domain (NOD) proteins that function in a manner similar to that of the TLRs in that they also recognize microbial components [5-7]. However, the initiating signals and downstream signaling pathways that are involved in the TLR and NLR systems are markedly different. TLRs mainly detect exracelluar PAMPs such as bacterial lipopolysaccharide (LPS) and bacterial flagellin, whereas NLR recognize peptidoglycans of bacteria [4,5,8]. LPS is an activator of the innate immune system against gram-negative bacterial infection, whereas peptidoglycans, which are the major component of gram-positive bacterial cell wall, are recognized by immune system against gram-positive bacterial infection [9,10]. NOD1, a member of the NOD proteins, is a cytosolic PRR that has an important role as an activator of the innate immune response in IECs. NOD1 is restricted to sensing γ-glutamyl diaminopimelic acid from gram-negative bacteria [11], whereas NOD2 is able to act as a general sensor of bacterial infection by recognizing peptidoglycans from both gram-positive and gram-negative bacteria [12,13].
Intercellular adhesion molecule-1 (ICAM-1, CD54) is a transmembrane glycoprotein that serves as a cognant-receptor for the β2-integrins (leukocyte functional antigen [LFA-1, CD11a/CD18] and [Mac-1, CD11b/CD18]) expressed on phagocytes [14]. ICAM-1 has a restricted tissue distribution and is constitutively expressed at low levels on subpopulations of hemopoietic cells, vascular endothelium, fibroblasts, and various epithelia such as bronchus, kidney, urinary tract, and skin [15]. Several human colon epithelial cell lines express ICAM-1, and its expression can be up-regulated by the cytokines TNF-α, IL-1β, and interferon γ (IFN-γ) and also by enteroinvasive bacterial LPS [16,17]. ICAM-1 has a major role in the trafficking of neutrophils from the blood into the site of inflammation. After this extravasation, neutrophils activate antimicrobial mechanisms through the release of neutrophil extracellular traps and phagocytosis [18]. ICAM-1 is an important member of the immunoglobulin superfamily of proteins, and it is centrally involved in trafficking of leukocytes to endothelial and epithelial barriers [19]. The function and the importance of leukocyte adhesion for the generation and maintenance of inflammation have been demonstrated in many animal experimental systems, such as in Icam-1 knockout mice [20]. Expression of ICAM-1 is up-regulated following infection with invasive bacteria on human colonic epithelial cell lines or on human enterocytes in an in vivo model system [17]. Furthermore, ICAM-1 was expressed on the apical side of polarized IECs, and its increased expression was accompanied by increased neutrophil adhesion to these cells. This adhesion may be necessary for reducing further invasion of the mucosa by invading pathogens [17]. Importantly, patients with leukocyte adhesion deficiency suffer from recurrent bacterial infections and impaired wound healing [21].
The previous reports have revealed that NOD1 might be a crucial sensor for certain enteroinvasive bacteria that avoid TLR signaling [5,6,22]. One study reported that Caco-2 cells expressing a dominant-negative (DN) form of NOD1 failed to activate NF-κB and, consequently, had deficient IL-8 production after infection with enteroinvasive E. coli [22]. Upon exposure to inflammatory cytokines, local expression of ICAM-1 is greatly induced in the infectious foci. Nevertheless, little is known about the relationship between NOD1 and ICAM-1 expression in invasive bacteria-infected human IECs.
We hypothesized that, in response to enteroinvasive gram-negative bacterial infection, ICAM-1 expression might be dependent on NOD1 through NF-κB activation. The aim of this study was to investigate whether NOD1 can up-regulate ICAM-1 expression via NF-κB activation in bacteria-infected IECs that avoid the TLR pathway.
METHODS
Cell lines and cell culture
The human colon epithelial cell lines Caco-2 and HT29, derived from a human colorectal adenocarcinoma provided by professor Hyun Chae Jung (Seoul National University), were grown in Dulbecco’s Modified Eagle Medium supplemented with 10% heat-inactivated fetal bovine serum [23]. The media were supplemented with 2 mM L-glutamine. Cells were maintained in 95% air and 5% CO2 at 37°C.
Bacteria, cytokines, and other reagents
The following bacteria were used in this study: enteroinvasive E. coli ATCC 43892 (serotype O29:NM) [22], Shigella flexneri (serotype B). S. flexneri isolated from fecal sample of the patient with diarrhea. Sample was inoculated on MacConkey agar and Salmonella-Shigella agar and incubated at 37°C. Colorless colonies were subcultured and identified using Vitek 2 system (bioMerieux Vitek, Marcy-l’Etoile, France) following the manufacturer’s recommendations. Serotyping was performed by slide agglutination using serotyping kit (Joong Kyeom, Ansan, Korea). The confirmed Shigella isolates were kept in brain heart infusion broth with 50% glycerol at –80°C for further study. IFN-γ and IL-8 obtained from Peprotech (Rocky Hill, NJ, USA) were used as positive controls. An anti-myc monoclonal antibody was obtained from Santa Cruz Biotechnology (Santa Cruz, CA, USA).
Plasmids and transfection
A DN NOD1 expression vector (pcDNA3-Nod1∆CARD-myc) with a deletion of the caspase activation and recruitment domain (CARD) and an empty vector control (pcDNA3) were provided by G. Nunez (University of Michigan) [24] and Sung Joong Kim (Seoul National University College of Dentistry), respectively. Cells were plated in 24-well dishes and were transfected with this plasmid DNA by using Lipofectamine Plus (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions.
Transfection and generation of stably transfected cell lines of Caco-2 cells and HT29 cells
Caco-2 cells and HT29 cells were transfected with pcDNA3-Nod1ΔCARD-myc (DN NOD1) or with pcDNA3 by using Lipofectamine Plus [22]. G418 (0.5 mg/mL)-resistant colonies were isolated by using glass cloning cylinders. The production of DN NOD1 in cells stably transfected with pcDNA3-Nod1∆CARD-myc was determined by immunoblotting with a monoclonal anti-myc antibody. The Caco-2 and HT29 cell lines that stably expressed DN NOD1 were designated Caco-2 cell lines transfected with dominant-negative vector (pcDNA3-Nod1ΔCARD-myc) (CDN1) and HT29 cell lines transfected with dominant-negative vector (pcDNA3-Nod1ΔCARD-myc) (HDN1), respectively. The Caco-2 and HT29 cell lines that stably expressed empty vector were designated Caco-2 cell lines transfected with empty vector (pcDNA3) (CEV1) and HT29 cell lines transfected with empty vector (pcDNA3) (HEV1), respectively.
Infection protocols
We selected the different cell lines for E. coli O29 or S. flexneri for better invasion to cell lines. Caco-2 cell lines were infected with E. coli O29, and HT29 cell lines with S. flexneri. Epithelial cells grown to confluence in 24-well, 6-well, or 10-cm plates were infected with enteroinvasive E. coli at a multiplicity of infection (MOI) of 100 or with S. flexneri at MOI 1000. The cells were incubated with bacteria for 1 hour, after which extracellular bacteria were removed by washing. The cells were incubated for additional periods of time in the presence of 50 µg/mL of gentamicin to kill the remaining extracellular bacteria but not the intracellular bacteria [22].
Reverse transcription polymerase chain reaction (RT-PCR) and real-time RT-PCR
Total cellular RNA was extracted with an RNeasy mini kit (Qiagen, Valencia, CA, USA) and treated with RNase-free DNase to remove any contaminating genomic DNA. For RT-PCR, 1 µg of total cellular RNA was reverse transcribed, and cDNA was amplified as described previously [23]. The ICAM-1 primers were sense primer 5’-GAT GCT GAC CCT GGA GAG CA-3’ and antisense primer 5’-AGC ACT TGC GGT CCA CGA TG-3’; this set of primers yielded a PCR product that was 409 bp long. The β-actin primers were sense primer 5’-TGA CGG GGT CAC CCA CAC TGT GCC CAT CTA-3’ and antisense primer 5’-CTA GAA GCA TTG CGG TGG ACG ATG GAG GG-3’, and the IL-8 primers were sense primer 5’-ATG ACT TCC AAG CTG GCC GTG GCT-3’ and antisense primer 5’-TCT CAG CCC TCT TCA AAA ACT TCT C-3’in conventional RT-PCR. The β-actin primers were sense primer 5’-AAG ATG ACC CAG ATC ATG TT-3’ and antisense primer 5’-GCG ACA TAG CAC AGC TTC T-3’, and the IL-8 primers were sense primer 5’-ACA TGA CTT CCA AGC TGG CC-3’ and antisense primer 5’-CAG AAA TCA GGA AGG CTG CC-3’ in real-time RT-PCR [23,25]. After a hot start, the amplification occurred in 35 cycles of 45 seconds of denaturation at 95°C and 2.5 minutes of annealing and extension at 60°C. The negative control reactions contained no added RNA in the RT reaction mixtures and no cDNA in the PCR amplification mixtures. For real-time PCR, 1 µL of cDNA was amplified by using an ABI Prism 7700 sequence detection system (PE Applied Biosystems, Foster City, CA, USA) with 2x SYBR Green master mix (PE Applied Biosystems) [26]. Amplification of the expected single PCR product was confirmed on a 1% agarose gel that was stained with ethidium bromide. The δ-δ crossing threshold method was used for analysis of real-time RT-PCR.
Cell lysates and immunoblotting
Cells were lysed in ice-cold lysis buffer (10 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% NP-40, 0.1% sodium dodecyl sulfate, 0.1% sodium deoxycholate, 1 mM ethylenediaminetetraacetic acid [EDTA], 10 µg/mL of leupeptin, 10 µg/mL of aprotinin, 1 mM phenylmethylsulfonyl fluoride, 1 mM Na+ vanadate, protease inhibitor cocktail set III [1:200; Calbiochem, La Jolla, CA, USA]) for 30 minutes. The cell lysates were sonicated and centrifuged, and the protein contents of the lysates were assayed using a Bio-Rad protein assay kit [22]. For immunoblots, cell lysates (7.5 µg of protein/lane) were electrophoresed on sodium dodecyl sulfate and 10% polyacrylamide gels and transferred to nitrocellulose membranes (Hybond ECL, Amersham Pharmacia Biotech, Piscataway, NJ, USA). The membranes were blocked with Tris-buffered saline (TBS; 50 mM Tris base [pH 7.2], 150 mM NaCl, 2.6 mM KCl) containing 5% dry milk and 0.1% Tween 20 and incubated overnight at 4°C with a mouse anti-myc monoclonal antibody in TBS containing 5% dry milk and 0.1% Tween 20. The blots were washed in TBS-Tween 20, incubated with horseradish peroxidase-conjugated sheep anti-mouse antibody (1:2,000) and visualized by enzyme chemiluminescence. Equal protein loading was verified after the blot was stripped and reprobed with an antibody to β-actin.
Nuclear extracts and electrophoretic mobility shift assay
Nuclear extracts were prepared as described in the previous study [27]. Ten micrograms of nuclear extract was equilibrated for 20 minutes with 25,000 cpm of (γ-32P) ATP-end-labeled NF-κB oligonucleotide in a binding buffer (10 mM Tris-Cl [pH 8.0], 75 mM KCl, 10% glycerol, 0.1 mM EDTA, 2.5 mM MgCl2, 1 µg of poly [dI/dC]). The sequence of the NF-κB consensus oligonucleotide from the ICAM-1 promoter was AGCTTGGAAATTCCGGAGC [28]. Bound and free DNA were resolved by electrophoresis through a 6% high-ionic-strength polyacrylamide gel containing 5% glycerol at 250 V in Tris-glycine-EDTA electrophoresis buffer [28]. NF-Y binding activity was used as a loading control.
Flow cytometric analyses
The cells were plated in 6-well plates (BD Falcon, BD Biosciences, Franklin Lakes, NJ, USA) at a density of 2.5 × 105 cell/well in a final volume of RPMI 1640 with or without G418 (0.4 mg/mL). The original medium was aspirated off, and fresh serum-free media was added to wells. Duplicate wells were treated with medium alone, IFN-γ (40 ng/mL) or S. flexneri for 18 hours. Monolayers of cells were detached by incubation with 0.25% Trypsin/EDTA, washed with phosphate-buffered saline, and incubated with the primary antibody anti-human CD54 (ICAM-1, BD pharmingen, San Diego, CA, USA) at 4°C for 1 hour in the dark. The cells were washed twice and incubated with an optimal concentration of R-Phycoerythrin (R-PE)-conjugated rat anti-mouse IgG1 (BD pharmingen) at 4°C for 30 minutes in dark. Following two washes, the cells were fixed with 1% paraformaldehyde, and ICAM-1 expression was analyzed using a FACStar (Becton-Dickinson, Mountain View, CA, USA). Negative controls were incubated with an isotype-matched control antibody. The data were analyzed using Cell Quest (Becton-Dickinson) software.
Statistics
All experiments were replicated three times with triplicate repeated measures. Statistical analysis was performed by Mann-Whitney U test using IBM SPSS Statistics version 19 Doctor’s Pack (IBM Co., Armonk, NY, USA). p values of < 0.05 were regarded as significant differences.
RESULTS
DN NOD1 inhibited ICAM-1 mRNA expression by enteroinvasive bacteria
Several proinflammatory genes, including the potent neutrophil chemoattractants IL-8 and ICAM-1, are up-regulated in invasive bacteria-infected IECs. The expression both of ICAM-1 and IL-8 were up-regulated in CEV1 infected with E. coli O29:NM from 4 to 9 hours in this experiment (Fig. 1). Next, we confirm the DN Nod1 expression in cell lysates from uninfected CEV1, CDN1, HEV1, and HDN1 using immunoblotting as we had demonstrated the expression level of NOD1 in the previous our experiment to indicate the knockdown efficacy [24]. DN Nod1 expression was observed only in CDN1 and HDN1 (bottom panel of Figs. 2B and 3B).
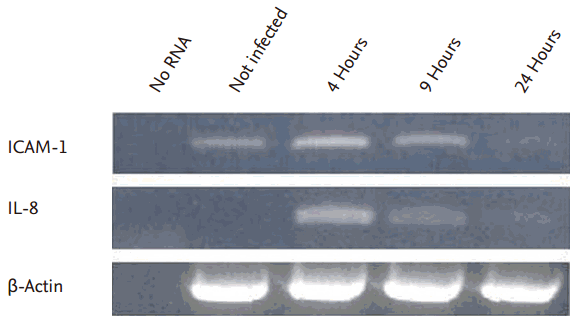
Intercellular adhesion molecule-1 (ICAM-1) expression in the Caco-2 cell lines transfected with empty vector (pcDNA3) (CEV1). ICAM-1 mRNA expression was up-regulated in CEV1 from 4 to 9 hours after infection with Escherichia coli O29:NM. IL-8, interleukin 8.
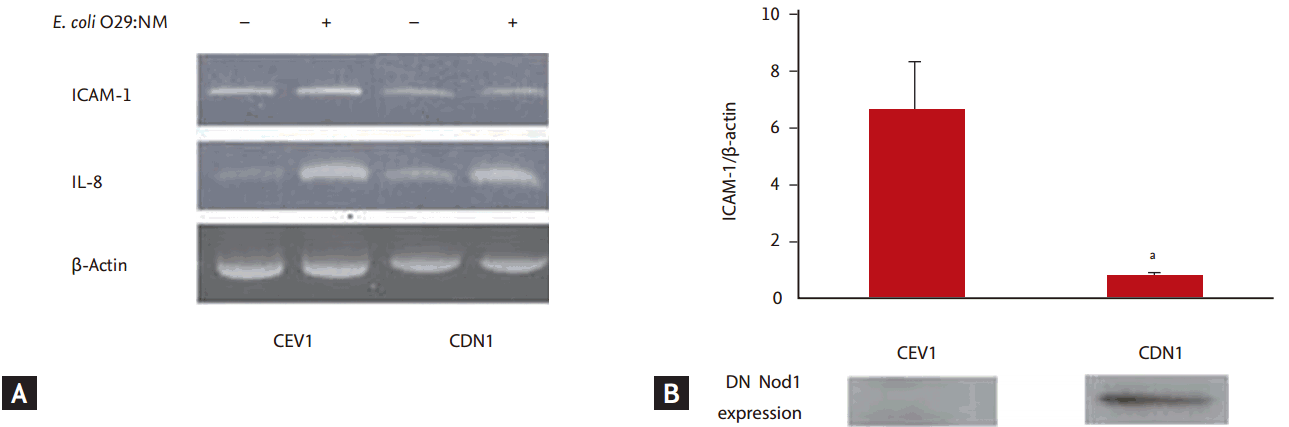
Intercellular adhesion molecule-1 (ICAM-1) mRNA expression in Caco-2 cell lines infected with Escherichia coli. (A) ICAM-1 mRNA expression was assessed by reverse transcription polymerase chain reaction (RT-PCR) 4 hours after E. coli O29:NM infection. ICAM-1 expression was inhibited in Caco-2 cell lines transfected with dominant-negative vector (pcDNA3-Nod1ΔCARD-myc) (CDN1) compared to Caco-2 cell lines transfected with empty vector (pcDNA3) (CEV1). (B) The amount of ICAM-1 mRNA assessed by real time RT-PCR was significantly reduced in CDN1 compared to CEV1. These results were from three independent experiments. The error bars represent standard error of the means. IL-8, interleukin 8; DN NOD1, dominant-negative nucleotide-binding oligomerization domain 1. ap < 0.05.
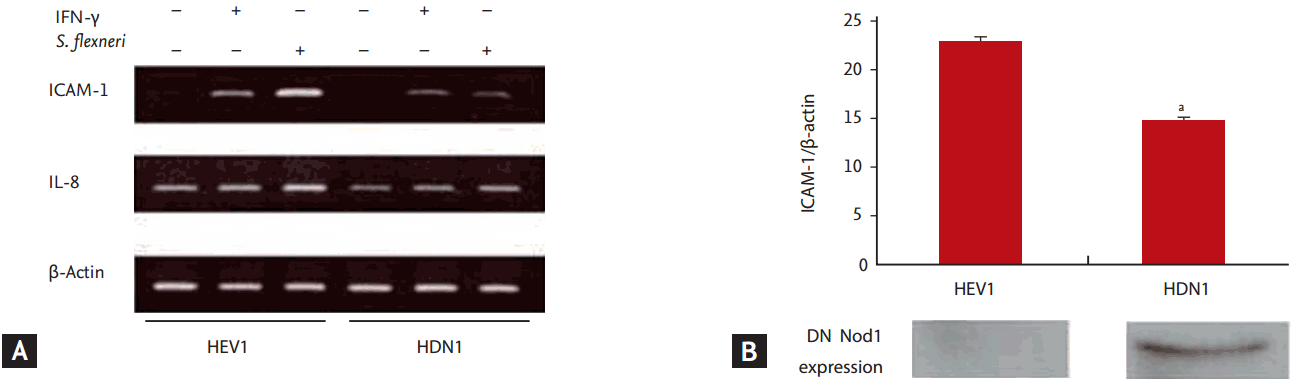
Intercellular adhesion molecule-1 (ICAM-1) mRNA expression in HT29 cell lines infected with Shigella flexneri. (A) The expression of ICAM-1 mRNA was assessed by reverse transcription polymerase chain reaction (RT-PCR) 4 hours after S. flexneri infection, and it was found to be inhibited in HT29 cell lines transfected with dominant-negative vector (pcDNA3-Nod1ΔCARD-myc) (HDN1) compared to HT29 cell lines transfected with empty vector (pcDNA3) (HEV1). (B) The amount of ICAM-1 mRNA assessed by real-time RT-PCR was significantly reduced in HDN1 compared to HEV1. These results were from three independent experiments. The error bars represent standard error of the means. IFN-γ, interferon γ; IL-8, interleukin 8; DN NOD1, dominant-negative nucleotide-binding oligomerization domain 1. ap < 0.05.
To determine whether NOD1 governs the expression of ICAM-1 in enteroinvasive E. coli-infected Caco-2 cells, CEV1 and CDN1 were infected with E. coli O29:NM, and then IL-8 and ICAM-1 mRNA levels were assayed by RT-PCR at 4 hours after E. coli O29:NM infection. DN NOD1 inhibited ICAM-1 mRNA expression in Caco-2 cell lines infected with E. coli O29:NM (Fig. 2A).
ICAM-1 mRNA quantity was assessed by real-time RT-PCR following E. coli O29:NM infection. The amount of ICAM-1 mRNA was significantly decreased in CDN1 compared to that in CEV1 (p < 0.05) (top panel of Fig. 2B).
Additionally, HEV1 and HDN1 were infected with S. flexneri. ICAM-1 mRNA expression was also found to be significantly decreased in HDN1 compared to that in HEV1, as measured by qualitative RT-PCR and real-time RT-PCR (p < 0.05) (Fig. 3).
Finally, cell surface expression of ICAM-1 was measured by flow cytometry, and parallel cultures were stimulated with IFN-γ. The number of cells that had higher fluorescence intensity was decreased in HDN1 compared to HEV1. This result means that cell surface expression of ICAM-1 increased in response to infection with S. flexneri infection in HEV1, but not in HDN1 (Fig. 4).
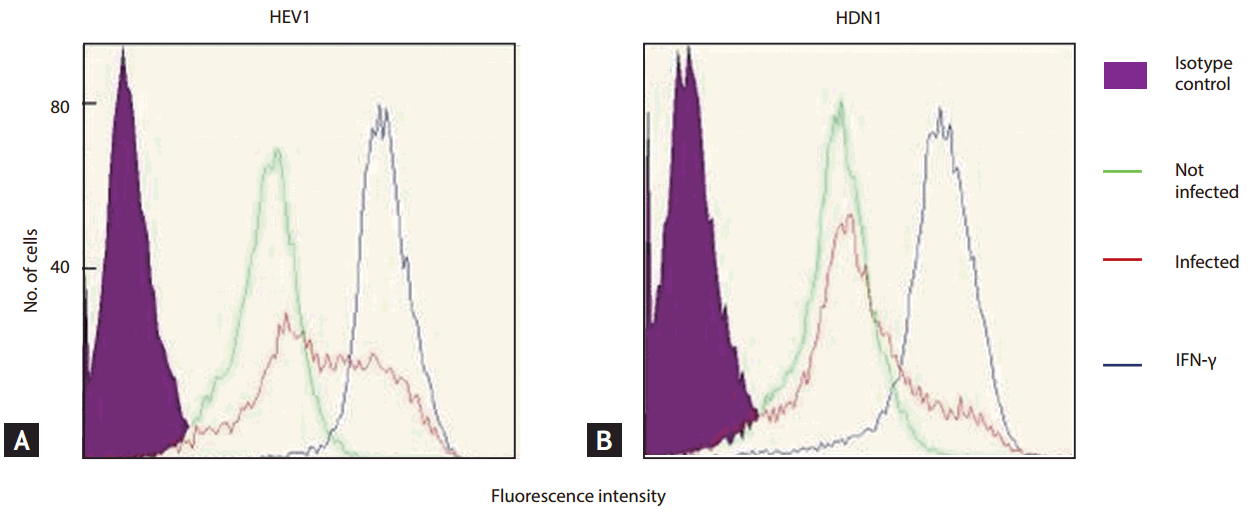
(A, B) Flow cytometric analysis of Intercellular adhesion molecule-1 expression in HT29 cell lines infected with Shigella flexneri. Parallel cultures were stimulated with interferon γ (IFN-γ). The number of cells that had higher fluorescence intensity was decreased in HT29 cell lines transfected with dominant-negative vector (pcDNA3-Nod1ΔCARD-myc) (HDN1) compared to HT29 cell lines transfected with empty vector (pcDNA3) (HEV1).
CDN1 inhibited NF-kB activation by E. coli O29:NM infection
Stimulation with IL-1α activates IKK and NF-κB in Caco-2 cells by a pathway that uses many of the same signal transduction intermediates as TLR5 [3,29]. Because activation of NF-κB through NOD1 is independent of TLR signaling pathways, we predicted that NF-κB activation in response to IL-1α would not be blocked in Caco-2 cells expressing DN NOD1. IL-1α activated NF-κB equally in both CDN1 and CEV1 in our experiment. However, as shown by electrophoretic mobility shift assay, CDN1 manifested marked inhibition of NF-kB activation in response to E. coli O29:NM infection when compared to CEV1 (Fig. 5).
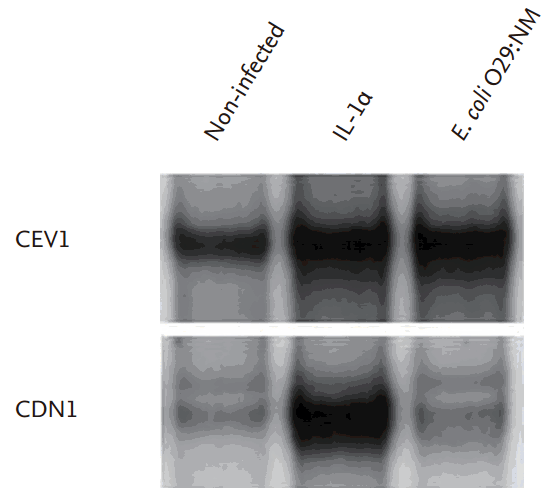
Nuclear factor kB (NF-kB) activation was assessed by electrophoretic mobility shift assay 45 minutes after infection or stimulation. NF-kB activation was inhibited in Escherichia coli infected Caco-2 cell lines transfected with dominant-negative vector (pcDNA3-Nod1ΔCARD-myc) (CDN1) compared with E. coli-infected Caco-2 cell lines transfected with empty vector (pcDNA3) (CEV1). However, dominant-negative nucleotide-binding oligomerization domain 1 (DN NOD1) did not eliminate NF-kB activation in response to interleukin 1 (IL-1).
DISCUSSION
Epithelial cells do not express TLRs on their apical surface and so are most likely unable to sense bacteria that are in the intestinal lumen [30]. In these cells, the NOD proteins are important activators of the innate immune response. These proteins are located in the cytosol of the cell and are divided into NOD1 and NOD2, which encode a CARD, either CARD4 or CARD15, respectively [11,31].
This study showed that ICAM-1 expression is dependent on NOD1 in human colon epithelial cells infected with a invasive gram-negative enteric pathogen that does not activate NF-κB through the TLR signaling pathway. This result implies that NOD1 plays an important role in the initial recognition of pathogenic bacteria at epithelial surfaces, such as the gut, where the innate immune responses to commensal bacteria must be avoided. Although human IECs and cultured cell lines such as Caco-2 also express other potentially functional TLR receptors (e.g., TLR3 and TLR9) [30,32,33], those TLRs, like TLR5, do not play a role in the activation of NF-κB by the enteroinvasive E. coli-infected colon epithelial cells [24,34].
DN NOD1 inhibited ICAM-1 mRNA expression in E. coli-infected Caco-2 cells and in HT29 cell lines infected with S. flexneri in this study. As measured by flow cytometry, ICAM-1 expression levels were minimally inhibited in CDN1 in response to infection of E. coli O29:NM (data were not shown), though DN NOD1 significantly inhibited the up-regulated expression of ICAM-1 in HT29 cells. These results are most likely due to high basal levels of surface ICAM-1 in Caco-2 cells [17]. In fact, ICAM-1 expression was observed in CEV1 without any inflammatory stimuli, as depicted in Figs. 1 and 2. However, as shown in Fig. 2, the quantitative real-time PCR measurement ICAM-1 expression of CDN1 decreased significantly compared to that of CEV1.
Microinjection of IECs with supernatants from S. flexneri (which contain the NOD1 ligand) activates translocation of the NF-κB subunit p65 to the nucleus in IECs from wild-type mice, but not in IECs from NOD1-deficient mice [5,35]. Because NF-κB can be activated through various other pathways, such as the TLRs, we used polarized human colon epithelial cell lines that are not responsive to bacterial LPS (TLR4) [36], but are responsive to signaling through TLR5 and other TLRs [27,32] to demonstrate the functional importance of signaling through NOD1 in the absence of other extracellular or intracellular PRRs [24]. Additionally, these cell lines were infected with wild-type pathogenic invasive Gram-negative bacteria that do not produce flagellin, which activates NF-κB by signaling through TLR5 [34]. As shown in Fig. 5, this study demonstrated that the activation of NF-kB by NOD1 was blocked, whereas signaling through IL-1R and TLRs was maintained. This finding supports our hypothesis that DN NOD1 significantly inhibits the activation of NF-κB in IECs following infection with enteroinvasive E. coli [24].
Human colonic epithelial cells are generally unresponsive to LPS. Therefore, colonic epithelial cells do not detect any potential threats from the normal bacterial flora. Detection of the resident bacterial flora by colonic epithelial cells would have serious consequences, as the colon would be in a state of chronic inflammation. However, these cells are not completely refractory to bacterial stimulation [37]. It has been suggested that colon epithelial cells are programmed to provide chemotactic and activating signals to adjacent and underlying immune and inflammatory cells in the initial period following microbial invasion. A specific array of four proinflammatory cytokines, IL-8, monocyte chemoattractant protein-1, TNF-α, and GM-CSF, was coordinately expressed and up-regulated in human colon epithelial cell lines in response to bacterial invasion or stimulation with TNF-α or IL-1 [23].
This study has limitations. First, we did not use the vascular endothelium. Instead, we used human colon epithelial cell lines. ICAM-1 is constitutively expressed at low levels on the surface of hemopoietic cells, the vascular endothelium, fibroblasts, and various epithelia, and its expression is usually increased in the blood vessel endothelium of the infection site [30]. Moreover, ICAM-1 is known to be expressed by several human colon epithelial cell lines and its expression can be up-regulated in these cells by enteroinvasive bacterial infection [5]. Therefore, our results can partially explain the ICAM-1 up-regulation mechanisms in human body. However, further study about ICAM-1 expression using NOD1 agonist [7] in vascular endothelium is warranted in the future to clarify the physiological role of NOD1 during bacterial infection. Nevertheless, this study provides information about the intestinal immune signaling pathway that may be helpful in the development of new antibiotic materials. Second, we did not show the linkage between NF-kB and ICAM-1 directly in this work by performing chromatic immunoprecipitation. However, taking into account the previous study reported that NF-kB was a central regulator of the IEC innate immune response including ICAM-1 activation, the linkage between NF-kB and ICAM-1 possibly had evidence [3]. Third, our results cannot be applied directly to all enteroinvasive bacterial infection mechanism. Further in vivo studies using various enteroinvasive bacteria are needed to conclude signaling pathway of enteroinvasive bacterial infection.
In conclusion, the present study demonstrated that NOD1 increases ICAM-1 expression through the activation of NF-kB in human IECs infected with E. coli O29:NM or S. flexneri. Signaling through NOD1 would play an essential role in neutrophil trafficking following infection with enteroinvasive bacteria.
KEY MESSAGE
1. Intercellular adhesion molecule-1 (ICAM-1) expression is dependent on nucleotide-binding oligomerization domain 1 (NOD1).
2. NOD1 increases ICAM-1 expression through the activation of nuclear factor kB.
3. NOD1 plays an important role in the epithelial recognition of pathogenic bacteria.
Notes
No potential conflict of interest relevant to this article was reported.