Bacterial Yield from Quantitative Cultures of Bronchoalveolar Lavage Fluid in Patients with Pneumonia on Antimicrobial Therapy
Article information
Abstract
Background/Aims
Early diagnosis and appropriate antimicrobial choice are crucial when managing pneumonia patients, and quantitative culture of bronchoalveolar lavage (BAL) fluid is considered a useful method for identifying pneumonia pathogens. We evaluated the quantitative yield of BAL fluid bacterial cultures in patients being treated with antimicrobials and attempted to identify factors predictive of positive BAL cultures.
Methods
Patients over 18 years old and whose BAL fluid was subjected to quantitative culture to identify the organism causative of pneumonia between January 1, 2005, and December 31, 2009, were included. We reviewed the results of BAL fluid bacterial cultures and the clinical records, laboratory tests, and radiographic findings of the patients.
Results
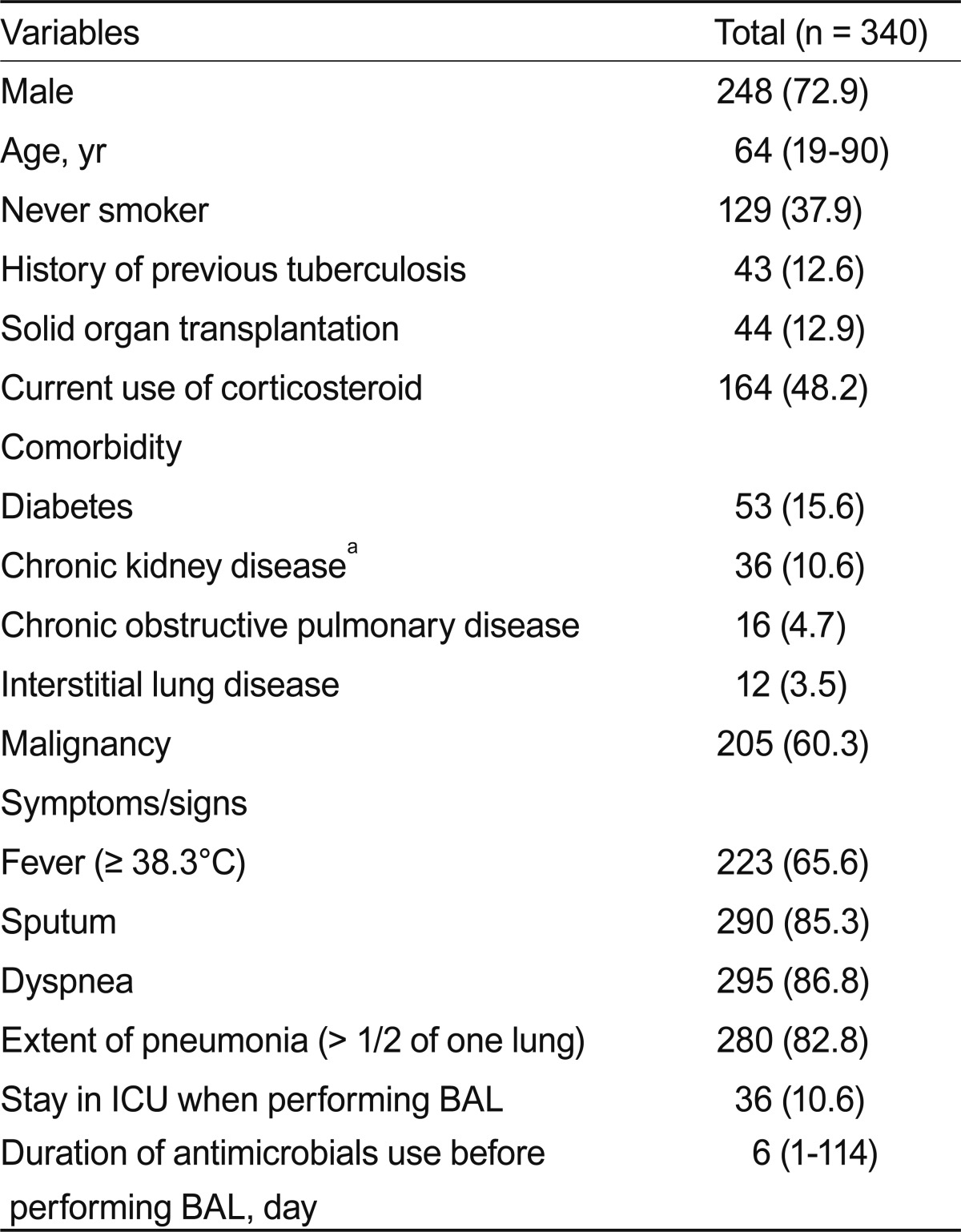
BAL was performed on 340 patients with pneumonia. A positive BAL culture, defined as isolation of more than 104 colony forming units/mL bacteria, was documented in 18 (5.29%) patients. Of these, 9 bacteria isolated from 10 patients were classified as probable pathogens. The most frequently isolated bacteria were methicillin-resistant Staphylococcus aureus, Acinetobacter baumannii, and Pseudomonas aeruginosa. No independent predictive factor for positive BAL cultures was identified.
Conclusions
The yield of quantitative BAL fluid bacterial culture in patients already on antimicrobials was low. Clinicians should be cautious when performing a BAL culture in patients with pneumonia who are already on antimicrobials.
INTRODUCTION
Pneumonia causes more deaths worldwide than any other infectious disease [1]. Although pneumonia can usually be cured with antimicrobials, patients can die if prompt, appropriate, and adequate therapy is not initiated [2]. Early diagnosis and proper choice of antimicrobials are crucial for successful management of pneumonia [3]. Bronchoalveolar lavage (BAL) and protected specimen brush are standard methods for identifying causative organisms [4,5]. The usefulness of endoscopic procedures, especially BAL, is well-established in both immunocompetent and immunocompromised patients with pneumonia [6-8]. BAL aids in identifying causative organisms in 50-70% of cases, which varies with both the type of pneumonia and the patient population [9-11]. However, previous antimicrobial therapy decreases the yield and accuracy of BAL for isolation of pneumonia-causing organisms. Unfortunately, BAL is usually performed in referral hospitals, where clinicians encounter patients with pneumonia who are already being treated with antimicrobials. The aim of this study was to determine the yield of quantitative BAL fluid bacterial culture in patients being treated with antimicrobial agents. We also attempted to identify factors predictive of positive BAL cultures.
METHODS
Study design and participants
Patients over 18 years of age in whom BAL through flexible bronchoscopy was performed to identify pneumonia-causing organisms from January 1, 2005, to December 31, 2009, at Seoul National University Hospital, a tertiary-referral university-affiliated hospital in Korea, were included in this retrospective cohort study. The protocol was approved by the ethics review committee of the hospital.
Review of clinical records
Patient demographics (gender, age, and smoking status) and clinical characteristics (comorbidities, symptoms, and physical signs) were obtained and reviewed. Results of laboratory tests and radiographic studies were also reviewed.
BAL procedure
Flexible bronchoscopy was performed using a standard protocol. Bronchoscopic examinations were performed by full-time faculty staff or fellows in the pulmonology division, and one or two well-trained nurses assisted the bronchoscopist with each procedure. A 5.9 mm diameter bronchoscope (model BF-1T240 or BF-1T260; Olympus Optical Co., Tokyo, Japan) was used. After advancing the bronchoscope through the vocal cords, 5 mL 1% lidocaine solution was infused through the scope, and additional volumes of 1% lidocaine solution were infused if needed during the procedure, with a maximum allowed dose of 5 mg/kg. After wedging the bronchoscope into subsegmental bronchi of the involved lesion, BAL was conducted using three or four 50 mL aliquots of sterile 0.9% warm saline infused through the working channel of the bronchoscope. Retrieved fluid was sent immediately to the Department of Clinical Microbiology for quantitative bacterial culture. BAL fluid was stained and cultured for aerobic and anaerobic bacteria, fungi (Aspergillus and Candida species), mycobacteria, and viruses. Bacteria present at a concentration of 1 × 103 colony forming units (CFU)/mL BAL specimen or higher were identified.
Definitions
Pneumonia
Pneumonia was defined as a new, progressive, or persistent (> 24 hours) infiltrate on a chest radiograph when two or more of the following criteria were met: macroscopic purulent sputum; temperature of > 38.5℃ or < 36.5℃; leukocytosis of 10,000 cells/µL or more, or leukopenia of less than 4,000 cells/µL; and dyspnea or worsening of respiratory status.
Positive quantitative bacterial culture from BAL fluid
A positive quantitative culture was defined when bacteria were cultured from BAL samples at a concentration of 1 × 104 CFU/mL or more.
Classification of isolates
True pathogens were defined as aerobic Gram-positive bacteria (Streptococcus pneumoniae, Staphylococcus aureus, and Group A hemolytic streptococci), aerobic Gram-negative bacteria (Klebsiella pneumoniae and Hemophilus influenzae), and atypical pathogens (Chlamydia pneumoniae or Mycoplasma pneumoniae), or an organism that could account for the clinical features of the patient. Probable colonizers or contaminants were defined as isolates obtained from BAL cultures that did not meet the criteria for a true pathogen.
Multidrug-resistant bacteria
Multidrug-resistant (MDR) bacteria included methicillin- resistant S. aureus, methicillin-resistant coagulase-negative staphylococci, vancomycin-resistant enterococci, S. pneumoniae resistant to penicillin and other broad-spectrum agents such as macrolides and fluoroquinolones, and MDR Gram-negative bacilli [12]. MDR Gram-negative bacilli included Klebsiella spp., Enterobacter cloacae, and Escherichia coli resistant to at least three of the following antimicrobial groups: third- or fourth-generation cephalosporins, aminoglycosides, fluoroquinolones, piperacillin, or ampicillin/sulbactam. Pseudomonas aeruginosa that was resistant to at least three of the following antimicrobial groups was also classified as an MDR Gram-negative bacillus: ceftazidime, cefepime, aminoglycosides, fluoroquinolones, carbapenems, and piperacillin. Acinetobacter baumannii, which is resistant to all antimicrobial agents, or all except imipenem, and organisms such as Stenotrophomonas maltophilia, which are intrinsically resistant to the broadest spectrum antimicrobial agents, were also considered MDR bacteria [12,13].
Treatment modification
Treatment modifications were defined as any change in antibiotic treatment, such as switching to another antibiotic, prolongation of antibiotic treatment, or supplementation of the preexisting antibiotic regimen.
Statistical analysis
Analyses were performed using Pearson's chi square test or Fisher's exact test for categorical variables, and Student's t test for continuous variables. The following variables were tested as possible predictors: extent of pneumonia (1/2 of lung), intensive care unit stay, duration of antimicrobial use before performing BAL, previous solid organ transplantation, and current use of corticosteroids. Variables with a p < 0.20 in univariate comparisons were subjected to multiple logistic regression to identify predictors of positive BAL cultures. Backward elimination was used in the logistic regression to select variables for the final model, and p < 0.05 was used as the criterion for significance of the associations. All statistical analyses were performed with SPSS version 17.0 (SPSS Inc., Chicago, IL, USA).
RESULTS
Baseline characteristics of patients
BAL was performed on 340 patients with pneumonia, comprising 248 men and 92 women with a median age of 64 years (range, 19 to 90). All patients were already on antimicrobials for treatment of pneumonia when BAL was performed. The median duration of antimicrobial use before performing BAL was 6 days (range, 1 to 114). Other demographic and clinical characteristics of the patients are shown in Table 1.
BAL quantitative culture
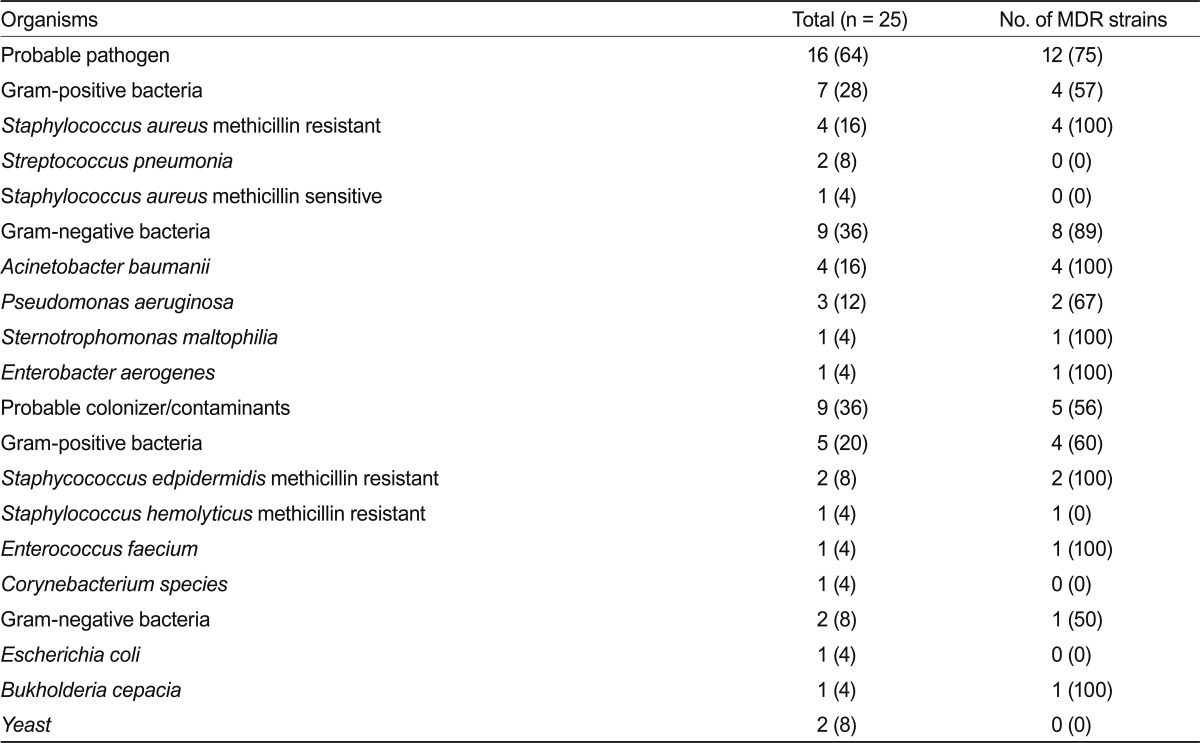
A positive BAL culture (> 10,000 colonies) was documented in 18 (5.29%) patients, and 25 microorganisms were isolated at concentrations above the diagnostic threshold. Among them, 9 probable pathogens were isolated from 10 patients (2.94%). A polymicrobial infection was present in three patients. The most frequently isolated species were methicillin-resistant S. aureus (four patients), A. baumannii (four patients), and P. aeruginosa (three patients) (Table 2).
Factors predictive of BAL culture-positivity
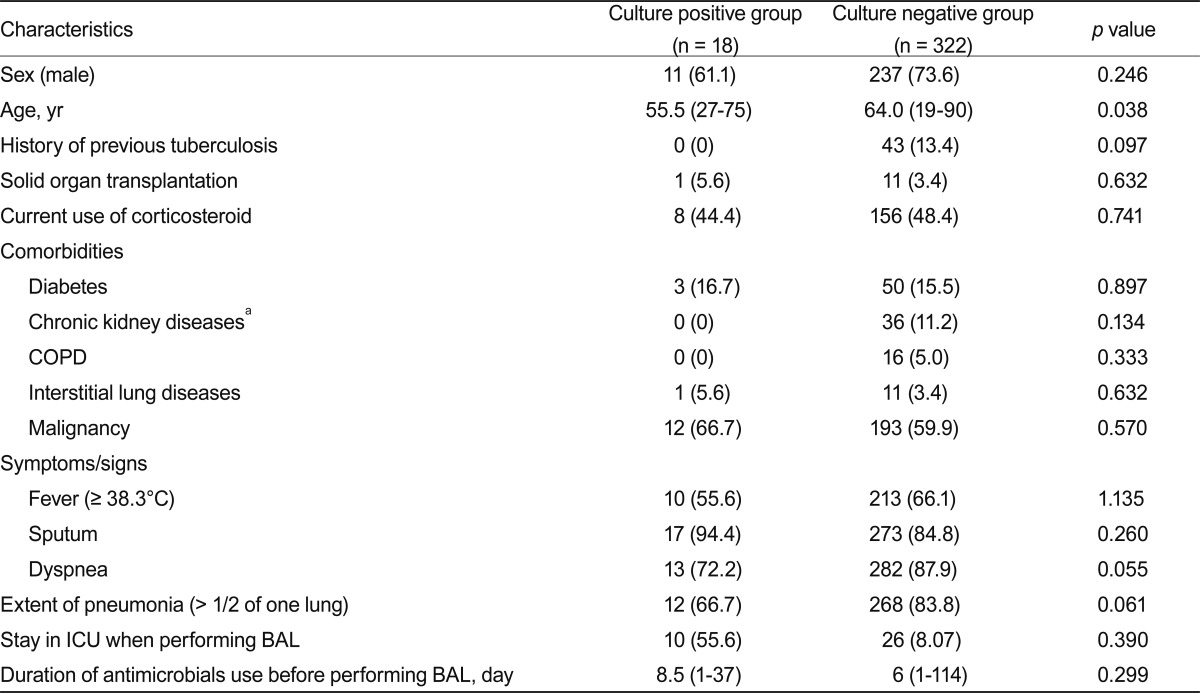
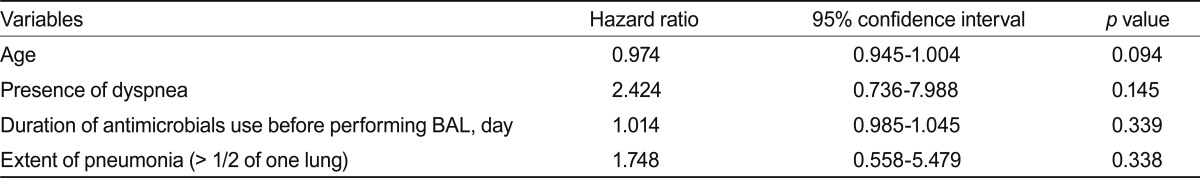
Patients with or without a cultured pathogen did not differ significantly with regard to comorbidities, clinical signs and symptoms, pneumonia extent, or duration of antimicrobial use before BAL (Table 3). Based on the clinical variables included in the univariate comparison of patients in the culture-positive and culture-negative groups, the final multiple logistic regression model predicting positive BAL cultures included age, presence of dyspnea, duration of antimicrobial use before BAL, and the extent of pneumonia. However, no independent predictive factor for positive BAL cultures was identified (Table 4).
Impact of BAL results on treatment
Treatment was modified in 6 of 10 patients with a probable pathogen identified in BAL fluid. Clinical improvement, such as a decreased need for a ventilator and a decreased extent of pulmonary infiltrates, was documented in four of six patients (67%).
DISCUSSION
Culture of BAL fluid was used to identify pneumonia-causing organisms. We found that the quantitative BAL fluid bacterial culture yield was as low as 2.94% in patients with pneumonia who were already on antimicrobials. This yield is much lower than that of previous studies, which reported 30-80% culture rates [14-17], possibly due to use of antimicrobials before BAL. Preculture antimicrobials significantly decrease the yield of blood and sputum cultures [18-21]. Furthermore, the long duration of antimicrobial treatment in our study (45% of patients received antimicrobials for more than 1 week before BAL) may have further decreased the yield. In a prior study, if BAL was performed within 3 days of starting antimicrobials, the overall yield was 63.4%, but decreased to 57.6% and 34.4% when performed within 3-14 days and after 14 days, respectively [14]. In addition, frequent use of wide-spectrum antimicrobials could decrease the yield of BAL fluid culture [22].
Despite the low BAL fluid culture yield, treatment was modified in 6 of 10 patients with a probable pathogen identified by BAL fluid culture; of these, 4 patients improved clinically. The other two patients from whom MDR pathogens (methicillin-resistant S. aureus and A. baumannii) were isolated deteriorated and died after a few days, even though pathogen-specific antimicrobials were administered immediately. Given that clinical improvement was achieved in four of six patients with a cultured pathogen, BAL culture is useful for determination of the appropriate antimicrobial therapy after isolation of a pathogen.
The majority of organisms cultured from BAL fluid were MDR bacteria (12 cases, 75%). The frequent isolation of MDR bacteria from our study population might be associated with the fact that our patient cohort had well-known risk factors for MDR bacteria, such as a nosocomial infection, admission within the previous 3 months, mechanical ventilation, indwelling central venous catheterization, and bladder catheterization [23].
Many previous studies focused on BAL quantitative cultures in patients with ventilator-associated pneumonia (VAP). However, we included not only VAP, but also community-acquired pneumonia and nursing-care-associated pneumonia to evaluate the overall yield of bacteria from BAL fluid cultures. Because the overall yield from quantitative BAL fluid cultures in patients on antimicrobials was very low, BAL performance before initiating antimicrobial treatment may be more appropriate, or its use should be delayed until prior antimicrobial therapy has proven ineffective.
The limitations of this study must be acknowledged. First, it was a retrospective design. Because the decision to perform BAL fluid quantitative culture depended on the individual clinician, confounding factors affecting the results could not be controlled for. In addition, we found no factors predictive of BAL culture-positivity. Positive cultures were not associated with the extent of pneumonia or duration of antimicrobial use before the procedure; this may have been due to the very low BAL yield.
In conclusion, the quantitative BAL fluid bacterial culture yield for identification of pathogens causing pneumonia in patients already on antimicrobials was very low. Clinicians should exercise caution when deciding whether to perform a BAL culture in patients with pneumonia who are already on antimicrobials.
Notes
No potential conflict of interest relevant to this article was reported.