Relationships Between the Expressions of CDX1 and CDX2 mRNA and Clinicopathologic Features in Colorectal Cancers
Article information
Abstract
Background
CDX1 and CDX2 are members of the caudal-type homeobox gene family and control the proliferation and differentiation of intestinal mucosal cells. Their expressions are commonly reduced in colorectal cancer, but reports about the relationships between their expressions and clinicopathologic features are rare. The aim of this study was to examine the expressions of CDX1 and CDX2 mRNAs in colorectal cancers and to assess the relationships between their expressions and clinicopathologic features.
Methods
CDX1 and CDX2 mRNA expressions were analyzed by real-time polymerase chain reaction in 48 colorectal cancers and in adjacent non-tumorous normal mucosal tissue.
Results
CDX1 and CDX2 mRNA expressions were significantly reduced in colorectal cancer tissues versus normal mucosal tissues (p=0.001, p=0.042, respectively). As compared with paired normal mucosal tissues, colorectal tissues showed reduced CDX1 mRNA expression in 64.6% (31/48) and reduced CDX2 mRNA expression in 66.7% (32/48) of cases. A statistically significant positive correlation was found between the expressions of CDX1 mRNA and CDX2 mRNA in colorectal cancer (r=0.543, p<0.001). However, the expressions of CDX1 and CDX2 mRNAs were not related to age, sex, cancer location, differentiation, lymphatic or vascular invasion, lymph node metastasis, stage or serum carcinoembryonic antigen level.
Conclusions
CDX1 and CDX2 mRNA expressions were found to be significantly reduced in colorectal cancers, but these expressional changes were not found to be related to clinicopathologic features.
INTRODUCTION
The Caudal-type homeobox gene family members CDX1 and CDX2 regulate transcription during the early differentiation period in the human intestine, and they are sparingly expressed, from the beginning of the duodenum to the epithelial cells of the small intestine and the large intestine under normal conditions1).
In intestinal mucosae CDX1 and CDX2 show different expressional patterns, CDX1 expression increases on approaching the distal part of the intestine, whereas CDX2 expression is most prominent in the proximal part of the large intestine and reduces on approaching the distal part of the large intestine2-5). Differences between the expressions of these genes according to the crypt/villus axis have been diversely reported by many studies2-4, 6, 7). CDX1 mRNA and protein are limited primarily to crypt cells, and although CDX2 mRNA is also detected in crypt cells, it is detected more so in differentiated cells moving toward the apex of villus2, 3, 6, 7). In the proximal colon, where the expression of CDX2 is most prominent, CDX2 mRNA and protein are detected evenly in crypt through to differentiated villus cells, and in the distal colon, CDX2 mRNA expression to crypt bottom, whereas CDX2 protein is expressed on cell surfaces4).
CDX1 plays important roles in the regulations of the proliferation and differentiation of intestinal epithelial cells6), whereas CDX2 has key roles in the transcriptional regulations of various genes related to intestinal epithelial cells and in the proliferation and differentiation of intestinal epithelial cells8). These roles of CDX1 and CDX2 in the proliferation and differentiation of intestinal epithelial cells suggests that they also have roles in the development of colon cancer.
The role of CDX1 in colon cancer is controversial. It has been reported that CDX1 mRNA attenuation with antisense in a colon cancer cell line (Caco-2) suppressed cell proliferation9), and that the overexpression of CDX1 in epithelial cells other than intestinal epithelial cells induced cell proliferation in vitro and enhanced tumorigenicity in xenografts10), which suggests that CDX1 has a carcinogenic effect. In fact, intestinal metaplasia has been reported to be associated with ectopic CDX1 expression in gastric and esophageal cancer6). However, recently, the loss of CDX1 in colon cancer was reported, which suggests that CDX1 is a tumor suppressor gene6, 11, 12).
On the other hand, CDX2 has been shown to act as a tumor-suppressor gene. In fact, mice heterozygous for CDX2 were found to develop multiple polyps in the small and large intestines, though this was most evident in the proximal large intestine13-15). The morphological characteristics of these polyps are diverse, and this has been speculated to be due to the complete loss of CDX2 expression and function, reprogramming of intestinal differentiation then follows, and as a result polyps are generated with an epithelial cell phenotype, i.e., resembling those of the esophagus and stomach, in the proximal area of the gastrointestinal tract14). In addition, in colon tumors, CDX2 expression was reported to be reduced in-line with tumor differentiation16).
Reports about the relationships between the expressions of CDX1 and CDX2 and the clinicopathologic features in colorectal cancers are rare. The aim of this study was to examine CDX1 mRNA and CDX2 mRNA expression in colorectal cancers and to assess the relationship between their expressions and the clinicopathologic features.
MATERIALS AND METHODS
Study population
This study was performed on 48 patients (males, 32; females, 16; mean age 61.4 years) diagnosed as having colorectal cancer by endoscopic biopsy or curative surgery between July 2002 and March 2003 at the Pusan National University Hospital, and of these 35 patients underwent surgery. Expressional patterns of CDX1 and CDX2 mRNAs were evaluated with respect to gender, age, tumor site, differentiation degree, the presence of blood vessel and lymphatic invasion, the presence of metastasis in lymph nodes, disease stage based on the Astler-Coller classification, and the level of carcinoembryonic antigen (CEA) prior to surgery.
The study was approved by the Ethics Committee of Pusan National University College of Medicine and informed consent was obtained from all patients.
Methods
1) Acquirement of cancer and normal mucosal tissues
In all patients, during endoscopic examination, biopsy samples were obtained from the colorectal cancer and from the nearby normal mucosal tissue, and stored at -70℃ until required.
2) Extraction of total RNA
To obtain total RNA, biopsy samples were added to 1 mL of easy-BLUETM (iNtRON, Seoul), homogenized, and incubated at room temperature for 5 minutes. 200 µL of chloroform was then added, mixed for 30 seconds, incubated at room temperature for 5 minutes, and centrifuged at 12,000 rpm for 15 minutes at 4℃. 400 µL supernatant was then transferred to a new tube, the same amount of isopropanol was added, mixed by shaking, and reacted by leaving at room temperature for 10 minutes. After this reaction, the mix was centrifuged at 12,000 rpm for 15 minutes 4℃, the supernatant discarded, and the RNA was obtained as a pellet. Pellets were washed twice by adding 1 mL of 75% ethanol, mixed 2-3 times, centrifuged at 10,000 rpm for 5 minutes 4℃, and dried at room temperature. They were then dissolved in 20 µL of diethyl pyrocarbonate-treated distilled water, optical densities were measured at 260 nm, and RNA concentrations were determined.
3) cDNA synthesis
A master mix was prepared by mixing 1X reaction buffer, 5 mM MgCl2, 0.8 mM dNTP, 40 units of RNase inhibitor, and 160 units of M-MLV reverse transcriptase, and kept in ice until required. 1 µg of total RNA and oligo-p (dT)15 primer were then mixed, incubated for 15 minutes at 65℃, and placed on ice. Final volumes were adjusted to 20 µL with the master mix, and the reaction was allowed to proceed at room temperature for 10 minutes, and at 42℃ for 1 hour. Subsequently, the mixture was incubated at 99℃ for 5 minutes to inactivate enzymes, incubated in ice for >5 minutes at least, and stored frozen at -70℃ until required.
4) Real-time polymerase chain reaction (RT-PCR)
(1) Preparation of primers and probes
To compare the amplification levels of the CDX1 and CDX2 genes, β-actin was used as a reference gene. Primers and probes were synthesized by TIM MOLBIOL (Berlin, Germany) (Table 1).
(2) Preparation of the titration standard graph
cDNA was synthesized from 1 ug of total RNA extracted from MCF7 cells, and by diluting at various concentrations, cDNA templates were prepared. Using β-actin as a primer, real-time PCR was performed, and a standard graph was obtained by the analysis of graph monitoring (Figure 1). Reproducibility was confirmed by repeating the experiments 2-3 times, and by preparing cDNA templates at different concentrations. Samples positive for β-actin, CDX1, or CDX2 PCR products were selected, and results are presented as arbitrary concentrations of 1 to 10 folds increase.

Real-time quantification PCR. (A) Analysis of the amplification plots for the standard concentrations. Ten-fold serial dilutions (a range of 1×100 to 1×104) of standard cDNA were amplified in a LightCycler, using a β-actin hybridization probe. (B) Standard curve; plot of the crossing point (cycle number) against input target quantity.
(3) PCR
PCR was performed using LightCycler-DNA Master Hybridization Probes kits (Roche, Mannheim, Germany). 10×LightCycler-DNA Master Hybridization Probes, 25 mM MgCl2, H2O, primers, and probes were added at the required concentrations and the master mixture was prepared. 18 µL of this master mixture was injected into a capillary, the standard diluted serially by 10 times, the cDNA of each sample, or a negative control containing 2 µL H2O, was injected, mixed lightly, and centrifuged for a few seconds. The capillary was attached to a LightCycler (Roche, Mannheim, Germany), opened the reaction program, and various parameters were entered. PCR cycling conditions used were denaturation at 95℃ for 10 seconds, annealing at 55℃ for 10 seconds, and elongation at 72℃ for 8 seconds.
(4) Quantitation
Simultaneously to the completion of PCR reaction, the crossing point of each reaction sample was analyzed by a LightCycler, and the relative concentration of each sample was calculated. The concentrations of CDX1 mRNA and CDX2 mRNA are presented as the concentration obtained by the crossing point of each sample divided by the concentrations β-actin mRNA.
Statistical analysis
Differences in the expressions of CDX1 and CDX2 mRNA in normal mucosal and colorectal cancer tissues were analyzed using the Wilcoxon signed rank test. The correlation between the expressions of CDX1 mRNA and CDX2 mRNA in colorectal cancer was analyzed using Spearman correlation coefficients, whereas the correlations between the expressions of CDX1 mRNA and CDX2 mRNA and clinicopathological factors were analyzed using the Chi-square test or Fisher's exact test. p values <0.05 were considered to be statistically significant, and all statistical analyses were performed using SPSS for Windows Ver. 10.0 (SPSS Inc. Chicago).
RESULTS
The expressions of CDX1 and CDX2 mRNA in colorectal cancer and in normal mucosa
In colorectal cancers, CDX1 mRNA expression was significantly lower than in normal tissues (0.043±0.599 vs. 0.110 ±0.182, p=0.001) (Figure 2A) and, similarly, CDX2 mRNA expression was also significantly decreased (0.265±0.234 vs. 0.360±0.362, p=0.042) (Figure 2B).

CDX1 mRNA levels (A) and CDX2 mRNA levels (B) in paired non-tumorous normal mucosal and colorectal cancer tissues. CDX1 mRNA levels and CDX2 mRNA levels were significantly decreased in colorectal cancers tissues (p=0.001, p=0.042, respectively).
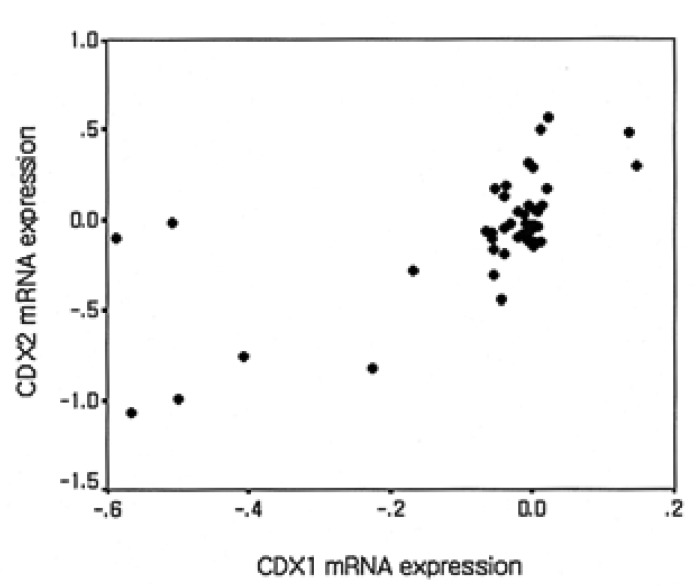
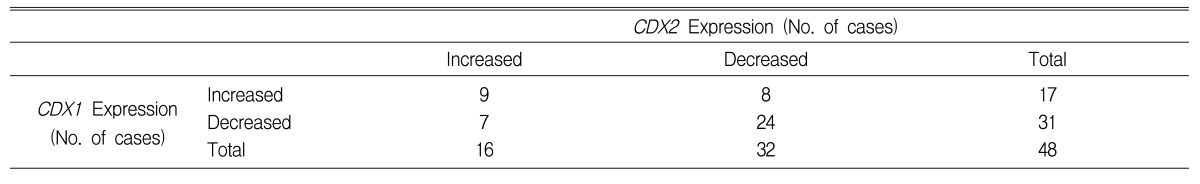
In colorectal cancers, the expressions of CDX1 mRNA or CDX2 mRNA were lower than in normal mucosal tissues in 64.6% (31/48) and 66.7% (32/48), respectively, and the expression of both CDX1 mRNA and CDX2 mRNA were decreased in 50.0% (24/48) of cases. And CDX1 mRNA and CDX2 mRNA expressions were both increased in 18.8% (9/48) of cases (Figure 3, Table 2). A statistically significant correlation was found between the expressions of CDX1 and CDX2 mRNAs (r=0.543, p<0.001) (Figure 4).

Comparison of CDX1 (A) and CDX2 mRNA levels (B) in colorectal cancer tissues and in paired normal mucosal tissues.
Relationship between the expressions of CDX1 mRNA and CDX2 mRNAs expression in colorectal cancers and compared with paired non-tumorous normal mucosasl tissues
Correlations between the expressions of CDX1 mRNA and CDX2 mRNA and clinicopathological factors
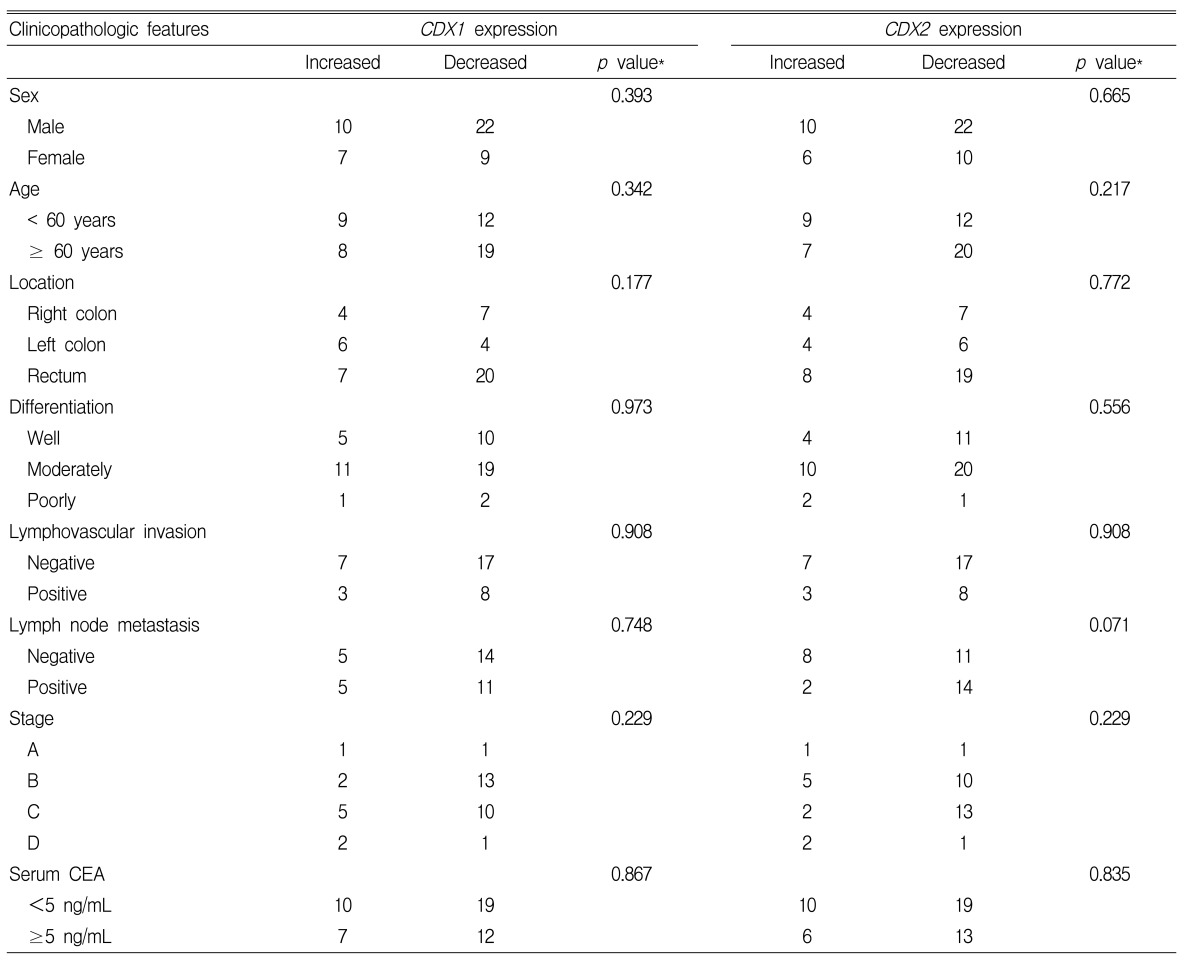
No difference was found between CDX1 or CDX2 mRNA expression and patient age (Table 3). The expression of CDX1 mRNA was reduced in 63.6% (7/11) of right colon cancers, in 40.0% (4/10) of left colon cancers, and in 74.1% (20/27) of rectal cancers, but no significant difference was found according to lesion site (p=0.177). The expression of CDX2 mRNA was reduced in 63.6% (7/11) of right colon cancers, in 60.0% (6/10) of left colon cancers, and in 70.4% (19/27) of rectal cancers, and similarly, no significant difference was found according to the lesion site (p=0.772) (Table 3).
Relationships between the expressions of CDX1 mRNA and CDX2 mRNA and clinicopathologic features in colorectal cancers patients
According to tumor differentiation levels, the expression of CDX1 mRNA was decreased in 66.7% (10/15) of well-differentiated types, in 63.3% (19/30) of moderately differentiation types, and in 66.7% (2/3) of poorly differentiated types. And, the expression of CDX2 mRNA was decreased in 73.3% (11/15) of well-differentiated types, in 66.7% (20/30) of moderately differentiated types, and in 33.3% (1/3) of poorly differentiated types. However, CDX1 mRNA and CDX2 mRNA expression levels were not significantly dependent on differentiation level (p=0.973, p=0.556, respectively) (Table 3).
The presence of lymphatic or vascular invasion was not found to be associated with the expression of CDX1 mRNA or CDX2 mRNA (p=0.908, p=0.908, respectively), and the presence of metastasis in lymph nodes was not associated with the expression of CDX1 mRNA (p=0.748). The frequency of CDX2 mRNA expression was lower in cases with lymph node metastasis than in those without; however, this was not statistically significant (57.9% vs 87.5%, p=0.071) (Table 3).
The expressions of CDX1 and CDX2 mRNAs were not significantly different at different disease stages (p=0.229, p=0.229, respectively) or on serum CEA level prior to surgery (p=0.867, p=0.835, respectively) (Table 3).
DISCUSSION
During the early developmental period of colon cancer, as has been observed in the normal distal intestine, incremental losses of CDX2 expression were detected, which became accentuated with cancer progression. In colon cancer, the expression of CDX2 decreases as dedifferentiation proceeds13, 16). In fact, it has been reported that the expression of CDX2 in adenoma with low-grade dysplasia and in adjacent normal tissues is similar, whereas the expression of CDX2 in adenoma with high-grade dysplasia is attenuated versus adjacent normal tissues16). In addition, numerous studies have reported reduced expressions of CDX1 and CDX2 in colon cancer6, 11, 12, 16).
In the present study, as compared with non-cancerous normal mucosa tissue, CDX1 mRNA and CDX2 mRNA were found to be reduced in 64.6% and 66.7% of tumor tissues, respectively, and 50% of cases showed concurrent reductions CDX1 and CDX2 mRNAs. This finding is in agreement with the report issued by Mallo et al.11), who found that in 12 cases of colon cancer CDX1 and CDX2 mRNA were either absent or reduced in 2 and 10 cases, respectively.
In colon cancer, few studies have been performed on the associations between the expressions CDX1 and CDX2 and pathologic differentiation. In colon cancer, because the expressions of CDX1 and CDX2 correlate inversely to dysplasia and prognosis, reductions in the expressions of CDX1 and CDX2 appear to be related to progression11). However, study, though the number of study subjects was small, i.e., 12 patients, we found that though the expression of CDX1 protein was reduced with progression in colon adenoma and colon cancer but that the pathological differentiation of colon cancer was not correlated with the expression of CDX1 protein, as determined immunohistochemically16). In the present study, the expressions of CDX1 and CDX2 mRNAs in colon cancer were not found to be related to the site of cancer development or differentiation level, or with gender, age, the presence of lymphatic or blood vessel invasion, the presence of lymph node metastasis, disease stage, or with CEA values prior to or after surgery.
We found a statistically significant correlation CDX1 mRNA and CDX2 mRNA expressions in colon cancers (r=0.543, p<0.001), which concurs with the findings of another study, which reported that during the development of colon cancer that CDX1 and CDX2 mRNA expressions are equivalently regulated11). This finding suggests the presence of a common mechanism underlying the expression of these two genes. However, it has also been reported that CDX1 mRNA and CDX2 mRNA are regulated via independent pathways11).
CDX1 and CDX2 are both required to maintain the differentiated phenotype of epithelial cells, and in poorly differentiated cells CDX1 and CDX2 were found to be mutated or down-regulated, a finding supported by a study which found that adenomatous polyps developed in mice heterozygous for CDX213-15).
In the normal colon epithelial cells of heterozygous CDX2 mice, CDX2 protein was found to be expressed, but it was not expressed in adenomatous polyps in these mice. This cannot be explained only by a loss of heterozygocity13), and it is not clear whether loss of CDX2 protein expression occurs secondary to mutation or whether it is simply due to a reduced level of normally differentiated gene products. Transcription of the CDX2 gene was found to be reduced in some colon cancer cell lines, in adenoma with high grade dysplasia, and in colon cancer; however, the presence of a mutation that inactivates the CDX2 gene in colon cancer has been reported to be extremely rare16-18). Similarly, reduced CDX1 expression in colon cancer may be due to a CDX1 gene alteration or a reduced level of tissue differentiation.
It may be speculated that during colon cancer development that reductions in the expressions of CDX1 and CDX2 may be involved in disease progression, alternatively, it may be speculated that the expressions of CDX1 and CDX2 are reduced due to a secondary change caused by colon cancer progression. The expressions of CDX1 and CDX2 are enhanced on moving from embryogenesis to intestinal epithelial cell maturity, and thus CDX2 could be of use as a marker of the normal epithelial cell phenotype. Hence, losses of CDX1 and CDX2 expression in colon cancer may be secondary changes associated with the loss of the normal intestinal epithelial cell phenotype.
In a previous study on CDX2 protein expression16), the expression of CDX2 protein was found to be decreased in most colon cancers. However, in one case strong CDX2 expression was detected. In our study of 48 cases, increased CDX1 mRNA and CDX2 mRNA expressions were detected in 17 cases and 16 cases, respectively, and this may be due to phenotypic differences in colon cancer19).
Most quantitative and qualitative studies on the expressions of homeobox genes have were speculated that these genes act as tumor suppressor genes if downregulated and act as tumorigenic genes if up-regulated. In our study, as the expressions of CDX1 mRNA and CDX2 mRNA were down-regulated in many colorectal cancers, they could be viewed as potential tumor suppressor genes. However, alternatively, this downregulation may be the result of a secondary change or the result of colon cancer phenotypic differences due to the loss of a normal intestinal epithelial cells phenotype due to cancer progression. We conclude that more study is required to determine whether CDX1 and CDX2 are directly related to the development of colon cancer.
In the present study, the expressions of CDX1 mRNA and CDX2 mRNA in colorectal cancer were found to be lower than in normal mucosa, but no correlation was found between their expressions and clinicopathological factors. However, since the expressions of CDX1 and CDX2 mRNAs were elevated in some colon cancers, we believe that more study of the roles of CDX1 and CDX2 during colon cancer progression is required.