Hepatitis B Virus DNA Detection by In Situ Hybridization in Human Hepatocellular Carcinoma
Article information
Abstract
The distribution of hepatitis B virus (HBV) DNA in tumor tissue sections from six Korean patients with HBsAg positive hepatocellular carcinoma (HCC) was examined by in situ hybridization using a biotin-labeled recombinant, cloned HBV DNA probe. All patients tested were positive for both HBeAg and anti-HBc in their sera. HBV DNA was distributed abundantly in the cytoplasm and rarely in the nuclei of tumor cells. The validity of the in situ hybridization assay was confirmed by the dot blotting technique using a 32P-labeled HBV DNA probe obtained by nick translation. In conclusion, it is speculated that integration of HBV DNA into host DNA as well as persistant amplified replication of the HBV DNA within the hepatocytes is linked etiologically to the development of human hepatocellular carcinoma.
INTRODUCTION
A revolutionary development in the field of molecular biology has been witnessed in the past decade in the various basic and applied sciences. This advance in recombinant DNA techniques permits the detection of hepatitis B virus (HBV) DNA by its ability to hybridize with radioactive or nonradioactive labeled HBV cloned DNA.1–5)
In addition to the epidemiologic evidences6,7) recent molecular biological investigations8–12) indicate a causal relationship between chronic infection of HBV and the development of hepatocellular carcinoma (HCC). The presence of HBV DNA, stably intergrated in the chromosomal DNA of infected human hepatocytes, in acute and chronic hepatitis, cirrhosis, and HCC has been reported.12–17)
The present study was undertaken to investigate the distribution of HBV DNA in Korean patients with HCC by the in situ hybridization technique using a biotinylated HBV DNA probe.
MATERIALS AND METHODS
Liver tissue samples from six patients with HCC were obtained by percutaneous needly biopsy under ultrasonographic guidance or by wedge biopsy at the time of surgery. The liver biopsies were fixed as for routine histologic analysis in 10% buffered formalin and embedded in paraffin blocks. For the in situ hybridization assay, microscopic slides and cover glasses were pretreated and subsequently acetylated as described by Haase et al.18) Five-micron-thick sections were floated at 45°C on sterile double distilled water (DDW) containing 0.5% (wt/vol) gelatin and 0.02% sodium azide. These sections were stretched on the pretreated clean slides. The slides were then baked 6 hours at 60°C. For dewaxing the slides were dipped into xylene twice for two 10-minutes each. Then after the sections were incubated for another 10-minutes in methanol containing 1.0% (vol/vol) H2O2 for inhibition of the endogeneous peroxidase activity. After washing in absolute enthanol for 10 minutes, the slides were hydrated by sequential 5-minute incubations in an ethanol-DDW mixture containing 95%, 80%, 60%, 30%, respectively. After washing in DDW for 5 minutes, the tissue sections were fixed in Carnoy’s A solution and dipped in 0.1% Triton X-100 in phosphate buffered saline solution (PBS), pH 7.2, for 2 minutes. The sections were washed in PBS and then placed in 0.2N HCl for 20 minutes to facilitate the penetration of the probe by diffusion.
Five minute after washing in DDW the slides were then placed at 70°C in 2× SSC (1 × SSC: 0.15M NaCl/0.015M Na citrate) and washed again in DDW for 5 minutes. The slides were then incubated for 15 minutes at 37°C with proteinase K (Sigma, St. Louis, MO.) (0.01 mg/ml in 20mM Tris-HCl, pH7.4, 2mM CaCl2) and rinsed in PBS containing 0.2% glycin to stop the proteinase K activity. After submersion in PBS containing 4% paraformaldehyde (Sigma, St. Louis, MO.) at room temperature for 20 minutes to stabilize the DNA,18,19) postfixation was carried out at 65°C for 15 minutes in 95% deionized formamide in 0.1 × SSC. The slides were then dipped for 2 minutes in a mixture of ice and 0.2 × SSC. Sequential dehydration was performed every 5 minutes through a graded ethanol-DDW series containing 50%, 70%, 80%, 90% and 100% ethanol and air dried briefly. Twenty μl of hybridization solution consisting of 10% dextran sulfate, 50% deionized formamide, 2mg/ml carrier DNA, 2 × SSC, and 5 μl/ml of the biotinylated HBV-DNA labeled by Bio-11-dUTP (ENZO Biochem, New York, NY.) through the nick translation as described by Rigby et al20) was applied to each slide. After covering each slide with pretreated clean coverglasses the hybridization reaction was adopted at 85°C for 10 minutes and then at room temperature for 30 minutes. The slides was then immersed in 2 × SSC and the coverslips were removed very carefully. The slides were incubated at room temperature with 50% deionized formamide in 0.1 × SSC for 10 minutes. The slides were when rinsed in a solution containing 0.05% Triton X-100 in 1 × PBS for 5 minutes at room temperature and finally in PBS for 3 minutes. In a humid chamber at room temperature the slides were covered with 100μl of the streptavidin-biotin-horseradish peroxidase complex (ENZO Biochem, New York, NY.) as described by the manufacturer. After tapping off the excess solution the slides were rinsed in 0.05% Triton X-100 in 1 × PBS. The slides were placed in the dark at room temperature for 20 minutes after applying 100μl of a solution containing 100μl of 1M Na acetate, 900μl DDW, 25μl of 1 % H2O2 and 20μl of aminoethylcarabazole (AEC) (ENZO Biochem, New, York, NY.) on the surface of section. After washing in DDW for one minute the slides were then counterstained with 0.25% (wt/vol) methyl green in 0.03M Na acetate, pH 4.8. Then after washing in 0.03M Na acetate, pH 4.8, for 2 minutes the slides were air dired briefly. After a dehydration in acetone-xylene (1:1 vol/vol) for 1 minutes and xylene alone for 3 minutes, the slides were mounted and examined by microscopy.
For confirmation of our findings, a dot blot assay of serum samples was also carried out by autoradiography of nitrocellulose filter paper using BRL hybridot-kit and 32P-labeled HBV-DNA probe (specific activity 106 cpm/μg DNA) kindly supplied by Dr. Roe, J. H. of the Department of Microbiology Seoul National University.
Serological markers of HBV (HBsAg, anti-HBs, anti-HBc, HBeAg, anti-HBe) were examined by radioimmunoassay using Abott kits (Abott Lab, North Chicago, IL.).
RESULTS
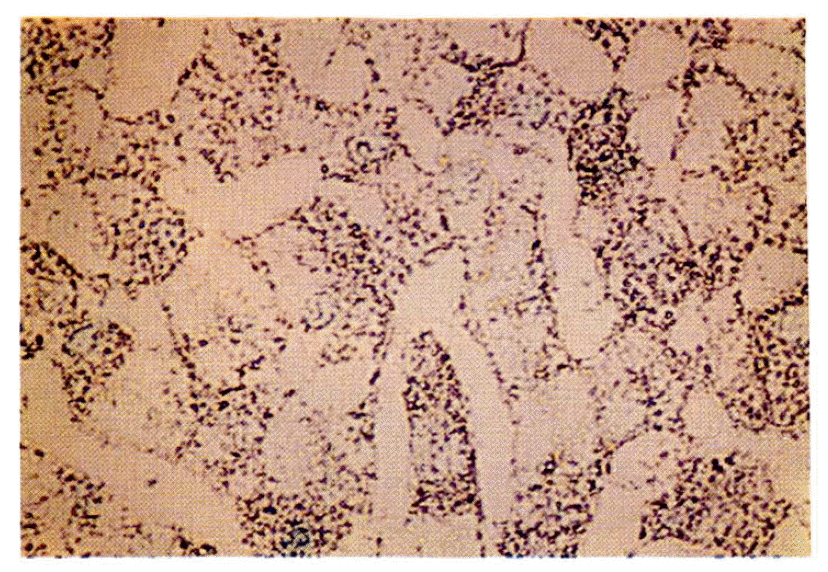
HBV DNA was found by in situ hybridization almost exclusively in the cytoplasm of HCC cells and infected hepatocytes of the cirrhotic area, in all six cases histologically diagnosed as HCC with liver cirrhosis. Contrarily, random distributions of HBV DNA in nuclei of hepatocytes were very rare (Fig. 1, 2, 3).

In situ hybridization of HBV DNA in hepatocellular carcinoma. The cytoplasm of HCC show abundant HBV DNA granules. The nuclei rarely reveal DNA grains. (Methyl green counterstain, × 400)

In situ hybridization of HBV DNA in the cirrhotic areas around HCC. The cytoplasm of the cirrhotic nodule shows plentiful HBV DNA grains. (×200)
Among all six cases, both HBsAg and HBeAg were positive in sera with positivity of anti-HBc.
By the dot blot analysis we detected HBV DNA at a lowest concentration of 40 pg/50μl in serum using a 32P-labeled HBV DNA probe (specific activity 106 cpm/μg DNA). This indicated, in turn, that we could confirm the validity of the in situ hybridization assay (Fig. 4).
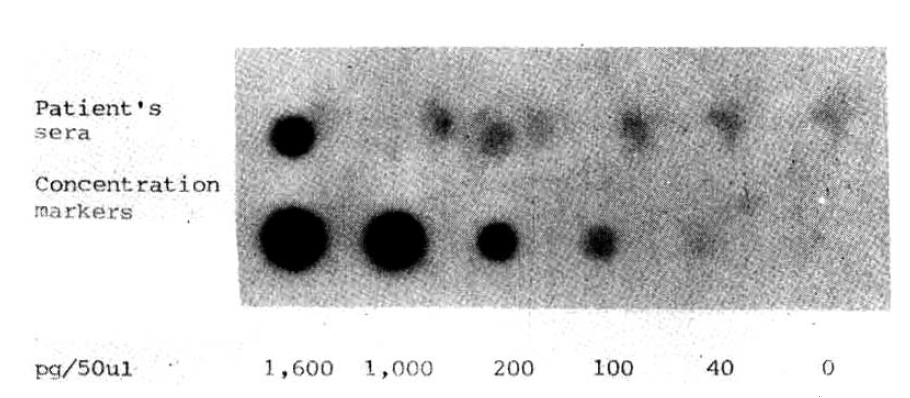
Autoradiography of the dot blot hybridization of sera of patients with HCC utilizing a32P-labeled HBV DNA probe (specific activity, 106 cpm/μg DNA). To determine the concentration, the indicated amounts of unlabeled recombinant lambda-HBV DNA with serial dilutions were directly spotted on the nitrocellulose paper in 50μl volumes, followed by hybridization with labeled DNA, and then autoradiographed for signal detection.
In the control liver tissue of a patient with fatty metamorphosis whose serologic markers for HBV were completely nil, there was no demonstrble HBV DNA by in situ hybridization.
DISSCUSSION
Immunohistochemically, HBV infection of chronic liver disease has been assessed by orcein staining,22) Victoria blue staining23) or peroxidase-antiperoxidase (PAP) staining.24) However, these are not always positive.23) In addition, they cannot analyze at the molecular level in a cell. Originally, in situ hybridization to determine genomic sequences on the tissue sections was described by Brahic and Haase25) using radioisotope-labeled probes which take 2–4 weeks to develop grains by autoradiography. Although this disadvantage has recently been overcome with the use of radioisotope-labeled probes of a relatively short functional half-life,19) radiation hazard remains to make it desirable to have an alternative approach.
The signal detection method of the avidin-biotin complex which uses biotinylated polynucleotides permits personal safety free from the isotope disposal problem, and sensitive and specific, and relatively rapid investigation.26) In addition, this means of HBV DNA detection in tissue sections does not require fresh-frozen tissue. Furthermore, a biotinylated DNA probe can be stored for a longer period than a radioisotope-labeled probe of short half-life. Again, the whole procedure can be carried out within 24 hours. Although dot or spot molecular hybridization is a suitable means to detect the free form of HBV DNA in serum, it cannot supply us with information of distribution in cellular structures in details.
In the present study, a pathologically nonspecific liver with mild fatty metamorphosis showed no HBV DNA by in situ hybridization. Contrarily, HBV genomic DNA were found in the tumor cells of all patients with HCC and serologically positive results of HBV infection. In view of the positivity of serological markers of HBV in all patients it is not surprising that we found plural distributions of free HBV DNA in the cytoplasm of malignant hepatocytes or infected hepatocytes of cirrhotic areas. This finding is consistent with previous reports.26,27) This means, in turn, amplification of rapid replication of HBV DNA in the cytoplasm of infected liver cells. Occasionally, the HBV, genomic DNA was found in the nuclei of hepatocytes. Regarding current reports11–16,28) for the integrated form of HBV DNA analyzed by Southern blot from tumor tissues in patients with HBsAg positive HCC, intranuclear HBV DNA may be related to the integration of the HBV DNA genome into the host cell DNA possibly manifesting a different expression of hepatocytes.29) Alternatively, this rare intranuclear distribution can be explained as a prerelease state of the HBV genomic DNA or conceivable as the formation of DNA-DNA and DNA-RNA hybrids. Generally, based on this finding it has been suggested that HBV is considered to be an oncogenic virus etiologically linked to the development of human HCC. Nevertheless, integrated HBV DNA has also been found in patients with various forms of chronic liver disease without evidence of HCC. Therefore the other aspects might be related to the liver carcinogenesis.
It has been recently stressed that the amplification of a protooncogene or oncogenes with qualitative and quantitative alterations is strongly concerned with multistep carcinogenesis.30–32) Recent reports32–34) indicated that the amplification of oncogenes occured from the G1-phase of the cell cycle to the mitotic phase during the regeneration of hepatocytes. In addition to this finding, enhanced oncogene expression in rodent liver carcinogenesis induced by application of a carcinogen on the regenerating states34,35) should be also kept in mind considering human hepatocarcinogenesis.36) It has been suggested that HBV may increase the expression of one or more protooncogenes by inserting itself in a critical regulatory position controlling the transcription of those protooncogenes.34) However, the precise role of HBV DNA in the amplification of oncogenes is still unknown.
Regeneration of hepatocytes by persistant HBV replication in an infected liver is presumably related to the triggering amplification of oncogenes and this, in turn, may occur in conjunction with a malignant transformation through a greater chance of mutation possible.
Acknowledgements
The authors thank Dr. Roe, J.H., Department of Microbiology, College of Natural Sciences, Seoul National University, for her help regarding the preparation of 32P-labeled HBV DNA probe by the nick translation procedure in dot blot hybridization.