 |
 |
| Korean J Intern Med > Volume 17(2); 2002 > Article |
|
Abstract
Background
Despite increasing importance of Acinetobacter baumannii in nosocomial infections and rapid development of multi-antimicrobial resistance in this strain, the resistance mechanisms of β-lactam antimicrobials in A. baumannii were not clearly defined. In order to observe the resistance mechanisms against β-lactams and carbapenem, we characterized the production of β-lactamases and outermembrane protein (OMP) profiles for the 44 clinical isolates of A. baumannii.
Methods
The MICs of antimicrobials were determined by agar dilution test. The secondary β-lactamases were characterized by isoelectric focusing, polymerase chain reactions and nucleotide sequencing, and the production of chromosomal β-lactamases was quantitated by spectrophotometric method. For two strains with an elevated MIC of carbapenem, outermembrane protein (OMP) profile was analyzed by ultracentrifugation of the sonicated bacteral cells and SDS-PAGE.
Results and conclusion
Twenty two or 4 of 44 strains produced TEM-1-like β-lactamase or PER-1 extended-spectrum β-lactamase, respectively. However, when we analyzed the MICs of several β-lactams with the β-lactamase production, the resistance level of β-lactam was mainly determined by the production of chromosomal β-lactamase, not by the secondary β-lactamases in the clinical isolates of A. baumannii. In two strains with an elevated MIC of imipenem, a decrease or loss of about 35 kDa and 22 kDa proteins in OMP was observed, which suggested that the change of OMP played a role in carbapenem resistance.
The widespread use of antimicrobials in modern hospitals results in the selection of the multidrug-resistant organisms. Acinetobacter baumannii have become one of the important emerging opportunistic pathogens because they are rapidly evolving toward multidrug-resistance and are often involved in various nosocomial infections that can be severe, such as bacteremia, meningitis, or pneumonia1). Because this organism is highly prevalent in nature, it is difficult to eradicate A. baumannii infection2). Despite this clinical importance, the pathogenesis and the mechanisms of antimicrobial resistance in A. baumannii are not clearly defined.
The β-lactam is one of the most commonly used antimicrobials in the world. In our hospital, β-lactam antimicrobials including carbapenem occupy more than 58% of all antimicrobials used. Although the resistance to β-lactam is rapidly increasing in A. baumannii, β-lactam antimicrobial is still a choice for the treatment of Acinetobacter infection3).
The objective of this study is to clarify the resistance mechanisms against β-lactams and carbapenem in clinical isolates of A. baumannii.
Forty four strains of A. baumannii were collected from Ulsan University Hospital and Dankook University Hospital during July, 1999. The strains were initially isolated and identified in the microbiology laboratories, and the isolates from different patients during the period were all included in this study. Strains carrying plasmids encoding β-lactamases TEM-1 (R1), TEM-3 (pCFF04), TEM-4 (pUD16), SHV-2 (pMG229), SHV-5 (pAFF2), CMY-1 (pMVP-1) served as the IEF standards4, 5), and P. aeruginosa carrying the genes encoding β-lactamases OXA-10 (R151), OXA-2 (pUD11) and Escherichia coli carrying PSE-1 (pMON811) served as the standards6–8).
Antimicrobials tested were piperacillin (Yuhan Co., Seoul, Korea); imipenem (Choongwae Pharma Co., Seoul, Korea); cefotaxime (Handok Pharmaceuticals Co., Seoul, Korea); ceftazidime (Glaxo Korea Co., Seoul, Korea); aztreonam (Dong-A Biotech Co., Seoul, Korea); clavulanic acid (Il-Sung Pharmaceuticals, Seoul, Korea); cloxacillin (Yuhan Co., Seoul, Korea).
MICs of antibiotics were determined by agar dilution method on Mueller-Hinton agar (Difco, Detroit, MI) with a Steers multiple inoculator according to the National Committee for Clinical Laboratory Standards9). E. coli ATCC 25922 and Pseudomonas aeruginosa ATCC 27853 were used as quality reference strains.
Crude preparations of β-lactamases from clinical isolates were obtained by sonication for 2×30 s at an amplitude of 12 μm in 0.1 M phosphate buffer (pH 7.0), with intermittent cooling on ice10).
Isoelectric focusing (IEF) was performed by the method of Matthew et al.10) by using an LKB Multiphor apparatus on prepared PAGplates (pH 3.5 to 9.5; Pharmacia Biotech Asia Pacific, Hong Kong) or Mini IEF cell system (Bio-Rad, Hercules, CA). Enzyme activities were detected by overlaying the gel with 0.5 mM nitrocefin in 0.1 M phosphate buffer, pH 7.0. Identification of β-lactamases was by comparison to reference enzymes, run in tracks adjacent to the test samples.
Inhibition assay was performed by overlaying the gels with 0.5 mM nitrocefin with and without 0.3 mM cloxacillin or 0.3 mM clavulanic acid in 0.1 M phosphate buffer, pH 7.011).
To amplify TEM-related genes from clinical isolates, the following oligonucleotide primers were synthesized and used for PCR; T1 5′-ATA AAA TTC TTG AAG ACG AAA-3′, T2 5′-GAC AGT TAC CAA TGC TTA ATC-3′. The PCR amplification was carried out as described elsewhere12). To amplify PSE-related genes from clinical isolates, the following oligonucleotide primers were synthesized and used for PCR: P1 (5′-TTA TTG GCA TTT TCG CTT TTA-3′), upstream 21 mer from the nucleotide 155; and P2 (5′-CGC ATC ATT TCG CTC TG-3′), downstream 17 mer from the nucleotide 942 of the PSE-gene, respectively11, 13). The PER-related β-lactamase gene was amplified by the primers PER1 and PER2; PER1 5′-ATG AAT GTC ATT ATA AAA GC-3′; PER2 5′-AAT TTG GGC TTA GGG CAA GAA A-3′. The PCR amplification was carried out as described elsewhere14). The PCR product of PER-1-related β-lactamase gene was directly sequenced. The nucleotide sequencing analyses were performed with BigDye™ Terminator Cycle Sequencing Ready Reaction kit and ABI 377 automated sequencer (Applied Biosystems Inc. Foster City, CA).
Cultures were grown overnight at 37°C in TSB, transferred with a 10% inoculum and incubated for 4 hours with shaking. The cells were subsequently harvested by centrifugation and resuspended in 1 mL of 0.1 M phosphate buffer, pH 7.0 and sonicated. The supernatants were assayed against 100 mM nitrocefin in 0.1 M phosphate buffer, pH 7.0 as substrate15). Activity was standardized for the protein concentration, assayed by the method of Lowry et al16).
The total membranes of the cells were isolated as described previously17) and electrophoresed using the 12% (wt/vol) sodium dodecyl sulfate-polyacrylamide gels. The 20 μg of OMP preparation was loaded in each well. The gel was stained with Coomasie Blue dye.
The MICs of piperacillin, cefotaxime, ceftazidime, aztreonam and imipenem were determined by agar dilution test (Table 1). Among 44 strains, 35 strains were highly resistant to piperacillin, cefotaxime, ceftazidime and aztreonam and their MIC50 values of piperacillin, cefotaxime, ceftazidime, aztreonam and imipenem were >256, >256, 256, 128 and 1 μg/mL. Seven strains were sensitive to piperacillin, cefotaxime, ceftazidime and aztreonam and their MIC50 values of piperacillin, cefotaxime, ceftazidime, aztreonam and imipenem were 16, 8, 4, 16 and 1 μg/mL, respectively. Two strains were sensitive to ceftazidime and aztreonam, but resistant to piperacillin and cefotaxime and their MIC50 values of piperacillin, cefotaxime, ceftazidime, aztreonam and imipenem were >256, 128, 4, 2 and 1 μg/mL, respectively.
Using the crude extracts of the 44 strains, we determined the pI values of β-lactamases produced by each strain. All of them produced the pI >8.0 chromosomal AmpC β-lactamases which were inhibited by 0.3 mM cloxacillin. Twenty two of 44 strains produced secondary β-lactamases with a pI 5.4, inhibited by 0.3 mM clavulanic acid, not by 0.3 mM cloxacillin. Four of 44 strains produced pI 5.3 β-lactamase which was inhibited by clavulanic acid but not by 0.3 mM cloxacillin. Because the β-lactamase with a pI 5.4, which showed clavulanic acid inhibition is mostly TEM-enzyme18), the genes of β-lactamase with a pI 5.4 were amplified with a TEM-specific PCR. All 22 strains were amplified with a TEM-specific PCR. Because pI 5.3 enzyme was inhibited by clavulanic acid but not by cloxacillin, which suggested the enzyme belonged to class A β-lactamase, we amplified the gene of pI 5.3 enzyme with TEM-specific, PSE-specific and PER-1-specific primers. The blagene was amplified with the PER-1-specific primers. The bagene of pI 5.3 enzyme was directly sequenced with the same primers for the PCR and the deduced amino acid sequence was identical with that of PER-1.
In order to find out the effects of the secondary β-lactamases to β-lactam resistance, we analyzed the MIC values of β-lactams according to the secondary β-lactamases which the strains produced (Table 2). The MIC50 values of piperacillin, cefotaxime, ceftazidime, aztreonam and imipenem in 22 strains producing pI 5.4 were >256, >256, 256, 128 and 1.5 μg/mL. Those values for the strains producing PER-1 were 96, >256, >256, >256 and 0.25 μg/mL and those for the strains which did not produce the secondary β-lactamases were 256, >256, 128, 128 and 1 μg/mL. These results cleary showed that pI 5.4 TEM-enzyme did not contribute the resistance against β-lactam antimicrobials in A. baumannii strains, but the pI 5.3 PER-1 enzyme seemed to raise the MIC of aztreonam.
Above data suggested that the resistance level of β-lactam antimicrobials in A. baumannii strains could be determined by other factors, such as the level of chromosomal β-lactamase production or membrane permeability. Thus, we quantitated the AmpC enzyme production by kinetic assay using several strains which were resistant or sensitive to all β-lactams and did not produce the secondary β-lactamases.
Among 18 strains which produced only the chromosomal AmpC β-lactamase, we selected 5 strains resistant to piperacillin, cefotaxime, ceftazidime and aztreonam and 3 strains sensitive to four β-lactam antimicrobials. The substrate was 100 mM nitrocefin and the kinetic study was performed in 0.1 M phosphate buffer (pH 7.0) at room temperature. The mean value of the β-lactamase production for the 5 strains which showed high resistance to four β-lactam was 254±130 U/mg protein and that for the 3 strains sensitive to β-lactam was 1.8±0.6 U/mg protein (1 Unit: 1 mmole nitrocefin hydrolysed/min at room temperature and pH 7.0) (Table 3). These data clearly showed that the level of the chromosomal AmpC β-lactamase production determined the β-lactam resistance in clinical isolates of A. baumannii.
Because the strains which showed an elevated MIC of imipenem did not produce a carbapenemase, we speculated that the porin changes in outermembrane protein (OMP) contributed to the imipenem resistance in A. buamannii. In order to observe the OMP, we isolated the OMP from two strains which showed a MIC of imipenem 2 μg/mL or above and three strains which are sensitive to imipenem and other β-lactams and analyzed by SDS-PAGE. The result of SDS-PAGE indicated the decrease or loss of about 35 kDa and 22 kDa protein in two strains which had an elevated MIC of imipenem (Figure 1). This validated our speculation; the loss or decreased amount of porin mediate the imipenem resistance in A. baumannii.
For the present, there were several β-lactamases identified which hydrolyzed carbapenem, such as IMP-1 in Japan and VIM enzymes in Europe and Korea19, 20). But the strains presented here did not produce the secondary β-lactamase which hydrolyzed carbapenem. Instead, OMP change contributed to the carbapenem resistance like other gram negative organisms, such as Pseudomonas aeruginosa21).
Twenty two and 4 of 44 strains produced the secondary β-lactamases such as TEM-1 and PER-1 respectively, but our results clearly indicated that the secondary β-lactamases did not have an important role in the resistance against the third generation cephalosporins in these clinical isolates of A. baumannii. In gram negative organisms which have an inducible chromosomal AmpC β-lactamase, such as Enterobacter cloacae and A.baumannii, chromosomal AmpC enzyme plays an important role in the resistance against the third generation cephalosporins. But our previous data showed that many of E cloacae strains resistant to the third generation cephalosporins did produce extended-spectrum β-lactamases (ESBL)22). Despite the fact that we did not quantitate the chromosomal AmpC enzyme production in the PER-1-producing strains, probably PER-1 ESBL mediated the resistance to extended-spectrum β-lactam antimicrobials in those strains. PER-1 ESBL in this study was identified for the first time in Korea.
In summary, the high percentage of clinical isolates of A. baumannii produced the secondary β-lactamases, mostly TEM-1, but the resistance against the third generation cephalosporin was mostly mediated by chromosomal AmpC b-lactamases. For the carbapenem resistance, the porin change in OMP played a role.
Acknowledgment
We are very grateful to Dr. Mina Kim for providing the clinical isolates of Acinetobacter baumannii.
Figure 1.
Comparison of the outermembrane proteins of the A. baumannii isolates (20 g of protein was loaded in each well): lane 1, molecular weight standards; lane 2 lane 4, the OMPs of the isolates sensitive to imipenem; lane 5 lane 6, the OMPs of the isolates resistant to imipenem. The arrows indicated decreased or loss of about 35 kDa and 22 kDa OMPs in lane 5 & 6
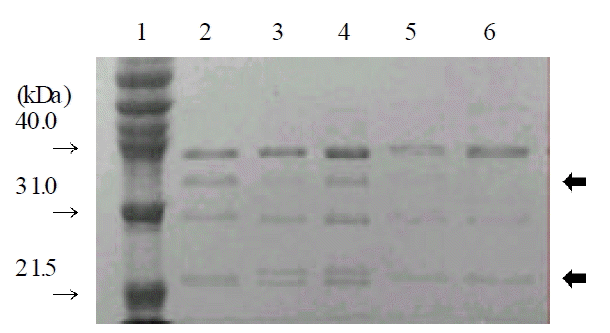
Table 1.
MIC (minimum inhibitory concentration) of several β-lactams for the 44 strains of Acinetobacter baumannii.
Table 2.
MIC (minimum inhibitory concentration) of several β-lactams for the 44 strains of Acinetobacter baumannii based on the secondary β-lactamase production.
Table 3.
MIC of β-lactams and chromosomal AmpC β-lactamase activity for the strains of A. baumannii.
| Strain No | MIC (μg/mL)1 | β-lactamase activity (U/mg protein)2 | ||||
|---|---|---|---|---|---|---|
| PIP | CTX | CAZ | AZT | IMP | ||
| Strains resistant to β-lactam antimicrobials | ||||||
| 7 | >256 | >256 | 256 | 128 | 2 | 202 |
| 8 | >256 | >256 | 64 | 128 | 2 | 483 |
| 15 | 256 | >256 | >256 | 128 | 1 | 173 |
| 16 | 256 | >256 | 256 | 266 | 1 | 181 |
| AR | >256 | >256 | 256 | 256 | 8 | 229 |
| mean±SD | 254±130 | |||||
|
|
||||||
| Strains sensitive to β-lactam antimicrobials | ||||||
| 21 | 16 | 8 | 2 | 8 | <0.5 | 1.1 |
| 33 | 8 | 4 | 4 | 8 | <0.5 | 2.1 |
| 34 | 8 | 4 | 4 | 8 | <0.5 | 2.3 |
| mean±SD | 1.8±0.6 | |||||
REFERENCES
1. Cohen ML. Antimicrobial resistance: prognosis for public health. Trends in Microbiol 2:422–4251994.

2. Villers D, Espaze E, Coste-Burel M, Giauffret F, Ninin E, Nicolas F, Richet H. Nosocomial Acinetobacter baumannii infections: microbiological and clinical epidemiology. Ann Intern Med 129:182–1891998.


3. Bergogne-Berezin E, Towner KJ. Acinetobacter species as nosocomial pathogens: microbiological, clinical, and epidemiological features. Clin Microbiol Rev 9:148–1651996.



4. Bauernfeind A, Chong Y, Schweighart S. Extended broad spectrum β-lactamase in Klebsiella pneumoniae including resistance to cephamycins. Infection 17:316–3211989.


5. Jacoby GA, Han P. Detection of extended-spectrum β-lactamases in clinical isolates of Klebsiella pneumoniae and Escherichia coli. J Clin Microbiol 34:908–9111996.



7. Jacoby GA, Medeiros AA. More extended-spectrum β-lactamases. Antimicrob. Agents Chemother 35:1697–17041991.



8. Jacoby GA, Han P. Detection of extended-spectrum β-lactamases in clinical isolates of Klebsiella pneumoniae and Escherichia coli. J Clin Microbiol 34:908–9111996.



9. National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. 3rd ed. Approved standard M7-A3. National Committee for Clinical Laboratory Standards, Villanova, Pa: 1993.
10. Matthew M, Harris M, Marshall J, Rose GW. The use of analytical isoelectric focusing for detection and identification of β-lactamases. J General Microbiol 88:169–1781975.

11. Danel F, Hall LMC, Gur D, Livermore DM. OXA-15, an extended-spectrum variant of OXA-2 β-lactamase, isolated from a Pseudomonas aeruginosa strain. Antimicrob. Agents Chemother 41:785–7901997.



12. Mabilat C, Goussard S. PCR detection and identification of genes for extended-spectrum β-lactamases. Diagnostic Molecular Biology Principles and Applications. In: Persing DH, ed. 553–559Mayo Foundation, Rochester, MN.: 1993.
13. Pai H, Kim JW, Kim J, Lee JH, Choe KW, Gotoh N. Carbapenem resistance mechanisms in Pseudomonas aeruginosa clinical isolates. Antimicrob Agents Chemother 45(2):480–4842001.



14. Vahaboglu H, Dodanli S, Eroglu C, et al. Characterization of multiple-antibioticresistant Salmonella typhimurium strains: Molecular epidemiology of PER-1-producing isolates and evidence for nosocomial plasmid exchange by a clone. J Clin Microbiol 34:2942–29461996.



15. Chen HY, Yuan M, Livermore DM. Mechanisms of resistance to β-lactam antibiotics amongst Pseudomonas aeruginosa isolates collected in the UK in 1993. J Med Microbiol 43:300–3091995.


16. Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem 193:265–2751951.


17. Poole K, Tetro K, Zhao Q, Neshat S, Heinrichs DE, Bianco N. Expression of the multidrug resistance operon mexA-mexB-oprM in Pseudomonas aeruginosa: mexR encodes a regulator of operon expression. Antimicrob. Agents Chemother 40:2021–20281996.



18. Pai H, Lyu S, Lee JH, Kim J, Kwon Y, Kim JW, Choe KW. Survey of extended-spectrum β-lactamases in clinical isolates of Escherichia coli and Klebsiella pneumoniae: Prevalence of TEM-52 in Korea. J Clin Microbiol 37:1758–631999.



19. Osano E, Arakawa Y, Wacharotayankun R, Ohta M, Horii T, Ito H, Yoshimura F, Kato N. Molecular characterization of an enterobacterial metallo beta-lactamase found in a clinical isolate of Serratia marcescens that shows imipenem resistance. Antimicrob. Agents Chemother 38:71–781994.



20. Yeom JH, Lee KS, Lee HM, Yong DE, Lee K, Kim JM, Chong Y. The characteristics of VIM-2 metallo-b-lactamase gene and their integron in clinical isolates of A. baumannii. Abstract no. 8 Korean Society for the Antimicrobial Chemotherapy 2001.
21. Trias J, Nikaido H. Outer membrane protein D2 catalyzes facilitated diffusion of carbapenem and penems through the outer membrane of Pseudomonas aeruginosa. Antimicrob. Agents Chemother 34:52–571990.



22. Pai H, Lee HJ. Personal communication.



 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Download Citation
Download Citation Print
Print



