 |
 |
| Korean J Intern Med > Volume 20(1); 2005 > Article |
|
Abstract
Background
To assess the possibility of VRE transmission from animals to humans, we studied the prevalence of vancomycin-resistant enterococci (VRE) in farm animals, raw chicken meat, and healthy people. We then determined the molecular relatedness of VRE isolates between animals and humans in Korea.
Methods
We aimed to isolate VRE from 150 enterococci specimens of farm animals, 15 raw chicken meat samples, and stools from 200 healthy people. Species differentiation was done with conventional biochemical tests. Vancomycin resistance genotyping was done by polymerase chain reaction (PCR). Using the agar dilution method, antimicrobial susceptibility was tested for 8 antimicrobials and pulsed-field gel electrophoresis (PFGE) was done to evaluate the molecular relatedness of VRE isolates.
Results
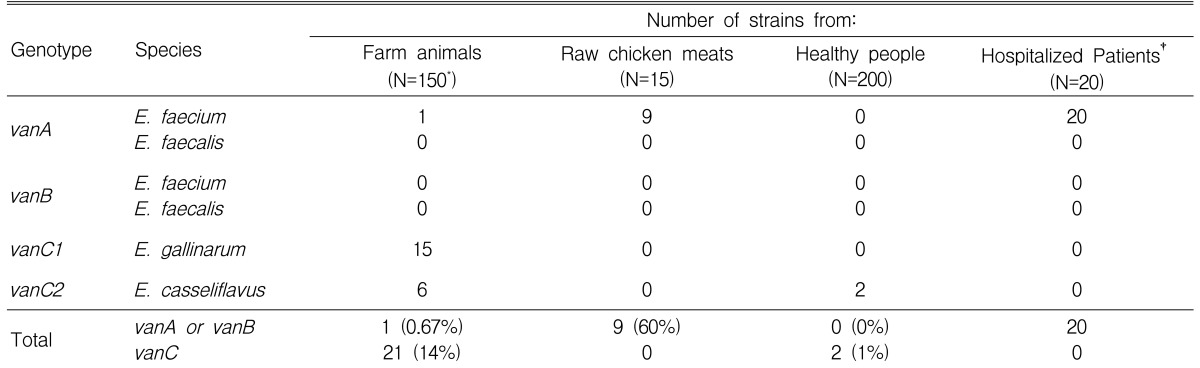
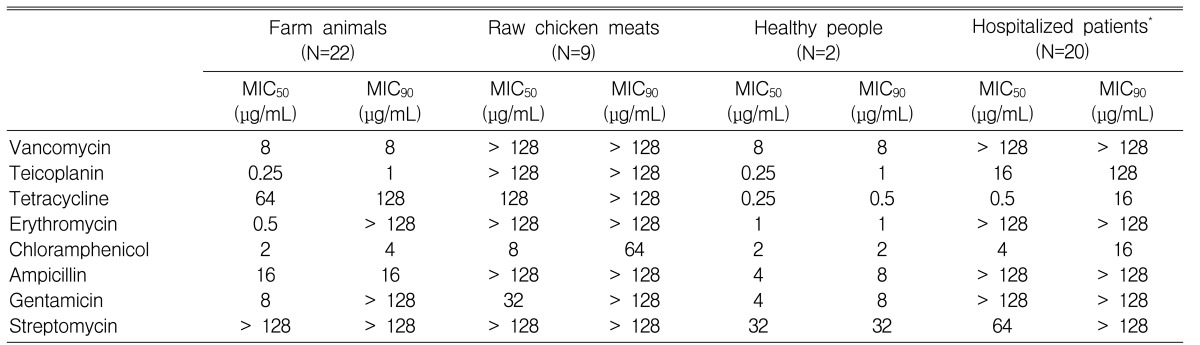
The prevalence of VRE was 14.7% (22/150) in farm animal specimens, 1% (2/200) in healthy people, and 60% (9/15) in raw chicken meat. Of 22 animal VRE isolates, 1 vanA E. faecium, 15 vanC1 E. gallinarum, and 6 vanC2 E. casseliflavus were identified. All of the 9 VRE from raw chicken meat and all of the 20 clinical VRE strains were vanA E. faecium. However, in healthy people, only 2 vanC2 E. casseliflavus were isolated. These showed low-level resistance to vancomycin and susceptibility to teicoplanin. However, 9 VRE strains from raw chicken meat had high-level resistance to vancomycin (MIC50,90: >128 ┬Ąg/mL), teicoplanin (MIC50,90: >128 ┬Ąg/mL), ampicillin (MIC50,90: >128 ┬Ąg/mL), erythromycin (MIC50,90: >128 ┬Ąg/mL), and tetracycline (MIC50,90: 128/>128 ┬Ąg/mL).
Conclusion
This study demonstrated little evidence of VRE colonization in healthy people despite high recovery
of VRE among raw chicken meat. It is suggested that there is little evidence of VRE transmission from animals to healthy people. However, we assumed that there exists the possibility of VRE contamination during the processing of chicken meat.
Enterococcus spp. is a facultatively anaerobic gram-positive coccus which is part of the normal intestinal flora. Some have also been isolated from the oral cavity and female reproductive tract. Enterococcus faecium colonizes in 10~25% of healthy individuals and Enterococcus faecalis exists in the gastrointestinal tract in almost 100% of people1). Enterococcus spp. has a relatively low virulence and does not easily induce disease in healthy individuals, but it causes a variety of opportunistic infections, such as bacteremia, urinary tract infection, endocarditis, and osteomyelitis. Because the use of antimicrobial agents, such as cephalosporin, in hospitals has rapidly increased since 1980, Enterococcus spp., which shows a relative resistance to these agents, has been highlighted as an important causative bacterium of nosocomial infections. It ranked second to E. coli as a nosocomial pathogen in the US in 19952). According to the National Nosocomial Infection Surveillance Study by the Korean Society of Nosocomical Infection Control in 1996, Enterococcus spp. accounts for 7.6% among the causative bacteria of nosocomial infections and ranks the fifth most frequently isolated pathogen. With the increasing isolation of Enterococcus spp., enterococci resistant to vancomycin, which has very effectively been used in treating Enterococcus spp. infection until recently, were reported in France for the first time in 19863). Since then, vancomycin-resistant enterococci (VRE) have continuously been on increasing trends in Europe and North America, and it was isolated domestically for the first time in 1992, showing a rapidly increasing trend centering around large hospitals since 199816, 18). The vancomycin resistance genotypes reported thus far include: vanA, vanB, vanC, vanD, vanE, and vanG, most of which are vanA and vanB types. They have clinical significance because they allow for plasmid-mediated transmission.
In the US, VRE infections are basically hospital-acquired infections and the prevalence of VRE among the general population is only 1%, whereas in Europe the subsisting rates of VRE are reported to be 0.5~14% in healthy individuals, and 12~28% in stock farm residents4, 17). The first susceptible route of VRE in healthy individuals is the oral administration of non-vancomycin-resistant Streptococcus faecium as a sort of bacterial treatment after being mixed with lactobacillus, which has an intrinsic vancomycin resistance in the intestinal tract, and subsequent resistance acquisition from it. The second susceptible route is contamination from food. In Europe, VRE was isolated when livestock were fed with avoparcin that had cross-resistance to vancomycin as a growth promoter, and since avoparcin was legally banned in 1997, VRE isolation rate has decreased4). Although it is also restricted domestically, its use was continued in stockbreeding. In 1998, its usage was banned. However, tylosin, bacitracin, and spiramycin, which can show cross-resistance to vancomycin, are still being used5). Domestically, Park et al. first reported the prevalence of VRE within poultry farms in 1999. According to their report, highly resistant VRE (vanA or vanB) were isolated in 2.58% of enterococcal isolates from chicken feces6).
According to the use of antibiotics and dietary habits of each country, various research results were shown. Although clinically significant factors such as the restriction on the use of antibiotics, the need for isolation, and the transmission of resistance are significant, domestic studies on the correlation between livestock isolates and clinical isolates, especially in healthy individuals, are insufficient. Therefore, we decided to begin by studying the colonization rate of VRE in livestock, raw chicken, and healthy individuals respectively, and secondly, to study the existence of cross infection between livestock and humans through antimicrobial susceptibility tests and molecular genetic studies.
The presence of VRE was looked for in the following sources: (i) 150 enterococci isolates from fecal samples of farm animals (114 chickens, 25 pigs, and 11 cows); (ii) intestinal secretions of 15 raw chicken meat samples that were purchased at 3 large dairy groceries (5 chickens for each grocery); (iii) fecal samples from 200 healthy people who had visited the Department of Health Examination in the Korea University Guro Hospital from March to May of 2002. VRE strains from each group were compared with 20 clinical strains that were available among 50 VRE isolates from hospitalized patients between January 2000 and December 2001.
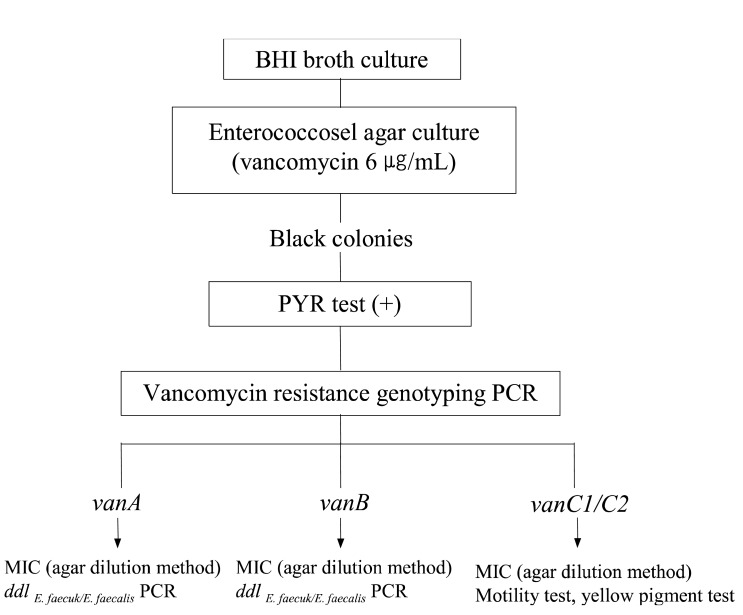
Each of the samples were suspended in BHI (brain heart infusion) broth (BBL, Becton Dickinson, USA) supplemented with 6 ┬Ąg/mL of vancomycin. After 16 hours of incubation, swabs were streaked onto enterococcosel agar (BBL, Becton Dickinson, USA) plates containing 6 ┬Ąg/mL of vancomycin if the broth was turbid. On the following day, black colonies from the enterococcosel agar plate were subcultured onto blood agar plates. Growth from the blood agar plate was used to test for and identify VRE. Colonies with the appearance of enterococci were tested for pyrrolidonyl arylamidase activity (PYR: Oxoid, England) to differentiate from lactococcus, pediococcus, and other bacteria. PYR-negative isolates were not investigated further14) (Figure 1).
Determination of glycopeptide resistance genotypes and confirmation of species identification were performed by polymerase chain reaction (PCR). The vancomycin resistance genes (vanA, vanB, vanC1, and vanC2) and ddl genes (E. faecium and E. faecalis) were amplified with the primers listed in Table 1. The PCR amplification mixture consisted of PCR buffer, 200 mM dNTPs, 25 pM primer, 1 U of Taq polymerase (BIOTOOLS, Spain), and DNA template. Reactions were set up in a final volume of 20 uL, containing 100 ng of boiled cellular supernatant and primer mixtures. Amplification conditions were 94Ōäā initially for 3 min; 94Ōäā for 1 min, 55Ōäā (50Ōäā for C2) for 30 s, 72Ōäā for 1 min over 30 cycles; and a final 10-min extension period at 72Ōäā. A reagent blank (containing all of the components of the reaction mixture except DNA) and positive control strains for each van genotype (E. faecium 1024 [vanA], E. faecium JB7 [vanB], E. gallinarum 11 [vanC1], E. casseliflavus ATCC 25788 [vanC2]) were run in every PCR procedure as control. DNA amplification was carried out with a PCR 2600 system (Perkin-Elmer Cetus, USA). Yellow pigmentation test and motility test was done for enterococcal isolates of vanC1 and vanC2 genotypes. For all of the VRE isolates, the ╬▓-lactamase stick (Oxoid, England) test was done.
MICs of vancomycin, teicoplanin, ampicillin, erythromycin, tetracycline, chloramphenicol, gentamicin, and streptomycin were determined by the agar dilution method following the National Committee for Clinical Laboratory Standards guidelines7). E. faecium JB7 was used as a reference strain. High-level resistance to aminoglycosides was determined by plating onto BHI agar containing 2,000 ┬Ąg of streptomycin per mL and BHI agar containing 500 ┬Ąg of gentamicin per mL. We analyzed the difference in the antimicrobial resistance rates of VRE isolates among farm animals, raw chicken meat samples, healthy people, and hospitalized patients.
For VRE strains, genotyping was done by PFGE. Overnight cultures grown on 5 mL of BHI broth were centrifuged for 1 min at 13,000 rpm. The pellet was suspended in 0.5 mL of 2X lysis buffer (12 mM Tris, 0.2 M EDTA, 2 M NaCl, 1.0% SLS, 1.0% deoxycholate) containing 1 mg/mL of lysozyme and 50 ┬Ąg/mL of RNase. An aliquot of this suspension (100 uL) was mixed with 100 uL of 1.6% agarose at 55Ōäā. This mixture was transferred into two 100 uL sample plug molds. The plugs were removed from the molds and were incubated for 18h at 37Ōäā in 3 mL of 1X lysis buffer (6 mM Tris, 0.1 M EDTA, 1 M NaCl, 0.5% SLS, 0.5% deoxycholate). This lysis solution was replaced by 3 mL of ESP buffer (10% SDS, 50 ┬Ąg/mL of proteinase K) and the mixture was incubated for 4 h at 55Ōäā. The plug was then washed for 1 h at 55Ōäā in 5 mL of diluted TE buffer (10 mM Tris, 0.1 mM EDTA) and the plug was washed again for 1 h with 20 mL of diluted TE buffer in a petri dish on the shaker. A slice of plug was digested with 20 U of the SmaI restriction enzyme (MBI Fermentas, Germany) and incubated for 18 h at 30Ōäā. DNA fragments were separated in a 1% agarose gel in 0.5X TBE buffer. Electrophoresis was performed with a CHEF-DR III apparatus (Bio-Rad, USA). Parameters for electrophoresis were 6 V/cm at 14Ōäā for 23 h, with pulse times ramped from 1s to 30 s. The gels were stained with 0.1 mg of ethidium bromide solution per liter for 30 min and were then placed onto a UV source. The size of the DNA fragments was then determined according to the size of the PFGE marker (Boehringer, Germany).
To assess the molecular relatedness of PFGE patterns of VRE isolates, the criteria of Tenover et al8) were used: (i) if an isolate differed from a main type by only three or fewer bands, it was considered to be a closely related subtype; (ii) if an isolate differed from a main type by 4~6 bands, it was considered to possibly be a related subtype; (iii) if an isolate differed from a main type by seven or more bands, it was considered to be a different type.
Comparison of the PFGE fingerprints was made using computer assisted analysis (Gel compar version 4.1: Kortrijk, Belgium). Comparisons of patterns were made using the unweighted pair group method using the arithmetic averages (UPGMA) clustering method by Dice coefficient. The band-width tolerance was set critically at 4.0%.
In the livestock, only 1 (0.67%) VRE isolate among the 150 enterococcal isolates was identified as vanA E. faecium. On the other hand, 21 (14%) isolates were identified to be vanC VRE, while 15 of them were E. gallinarum and 6 were E. casseliflavus. VRE was cultivated in 18 out of 114 chicken fecal samples (16%: 1 isolate of E. faecium, 15 isolates of E. gallinarum, and 2 isolates of E. casseliflavus), in 1 out of 25 pig fecal samples (4%: 3 isolates of E. casseliflavus), and 3 out of 11 cow fecal samples (27%: 3 isolates of E. casseliflavus).
In the samples obtained from the raw chicken meat, 11 out of 15 showed black colonies when cultivated in the enterococcosel agar, but 9 (60%) of them were PYR-positive and the 9 isolates were all identified to be E. faecium.
Among the 200 fecal samples of healthy individuals, 82 of them showed black colonies when cultivated in the enterococcosel agar. However, only 2 (1%) of them were PYR-positive and all were identified as E. casseliflavus. Clinically isolated VRE was vanA
E. faecium in all 20 isolates, and E. faecalis was not isolated (Table 2).
According to vancomycin resistance genotyping using PCR, the 22 livestock isolates included 1 vanA isolate, 15 vanC1 isolates, and 6 vanC2 isolates, and the 9 isolates from the raw chicken meat were all vanA type. The 2 E. casseliflavus isolates from healthy individuals were all vanC2 genotype, but the 20 clinical VRE isolates were all of the vanA genotype. The ddl PCR results for vanA VRE strains showed E. faecium in 1 livestock (chicken), 9 raw chicken meat samples, and 20 clinical isolates (Table 2).
According to the ╬▓-lactamase test, only 2 isolates (3.8%) out of the entire 53 VRE isolates were ╬▓-lactamase positive and all of them were E. faecium isolates from raw chicken meat.
The 22 VRE isolates from the livestock showed a low degree of resistance (MIC50,90: 8 ┬Ąg/mL) to vancomycin, susceptibility to teicoplanin (MIC50/90: 0.25/1 ┬Ąg/mL), and susceptibility or a low degree of resistance to ampicillin, erythromycin, and chloramphenicol but not tetracycline (MIC50/90: 64/128 ┬Ąg/mL), and streptomycin (MIC50/90: >128/>128 ┬Ąg/mL). All of the 9 isolates from raw chicken meat were vanA E. faecium, and also showed resistance to most of the antibiotics, including vancomycin (MIC50/90: >128/>128 ┬Ąg/mL) and teicoplanin (MIC50/90: >128/>128 ┬Ąg/mL)(Table 3).
The 2 vanC2 E. casseliflavus isolates from healthy individuals showed a low degree of resistance (MIC50/90: 8 ┬Ąg/mL) to vancomycin and susceptibility to teicoplanin (MIC50/90: 0.25/1 ┬Ąg/mL), while showing susceptibility or a low degree of resistance to ampicillin (MIC50/90: 4/8 ┬Ąg/mL) and tetracycline (MIC50/90: 0.25/0.5 ┬Ąg/mL), erythromycin (MIC50/90: 1/1 ┬Ąg/mL), chloramphenicol (MIC50/90: 2/2 ┬Ąg/mL), gentamicin (MIC50/90: 4/8 ┬Ąg/mL), and streptomycin (MIC50,90: 32 ┬Ąg/mL). All of the 20 clinical isolates were vanA E. faecium and showed resistance to most of the antibiotics including vancomycin (MIC50/90: >128/>128 ┬Ąg/mL) and teicoplanin (MIC50/90: 16/128 ┬Ąg/mL).
Interestingly, VRE isolates from farm animals or raw chicken meat showed a high resistance to tetracycline, but the isolates from healthy individuals and clinical isolates all showed susceptibility to this antimicrobial. Also, the raw chicken meat, livestock, healthy individuals, and clinical isolates all showed susceptibility to chloramphenicol, except the 2 raw chicken meat isolates.
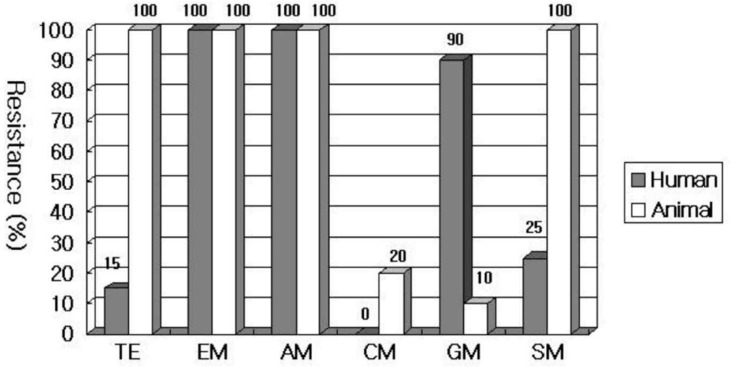
The vanA E. faecium isolates from the humans (20 clinical isolates) and the ones from the animals (9 raw chicken isolates and 1 livestock isolate) were compared in terms of the differences in their susceptibility to antibiotics. Erythromycin and ampicillin showed a 100% resistance in both humans and animals, but chloramphenicol showed a very low resistance rate, and all showed susceptibility in humans (humans/animals, 0/20% p=0.038). Tetracycline and streptomycin each showed a 100% resistance in the animals, but the resistance was relatively low in humans (tetracycline 15% p=<0.001, streptomycin 25% p=<0.001), and gentamicin showed a rather higher resistance rate in humans (p=<0.001) (Figure 2).
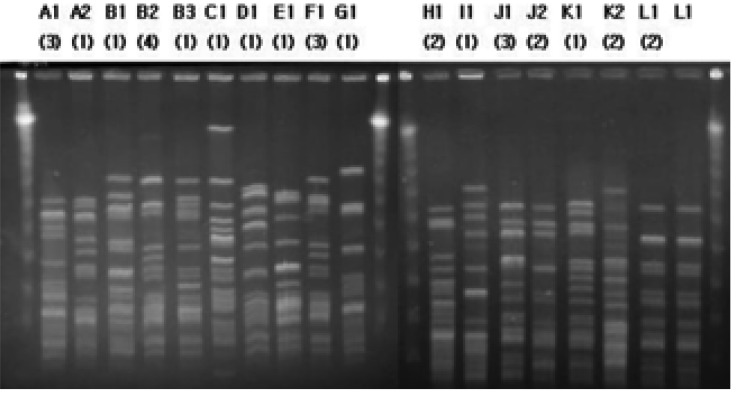
When the patterns of PFGE of vanA isolates were analyzed by the criteria of Tenover et al8), they were isolated in 12 types (A-L). For the clinical isolates, the PFGE patterns of the 20 isolates were manifested in 9 different types (A-I), and one type did not appear noticeably frequently. For the raw chicken meat isolates, 5 J-type isolates, 3 K-type or its subtype isolates, and 1 L-type were shown. The livestock vanA isolates did not show molecular types or the subtypes as the clinical isolates did, but showed molecular types such as the L-type of the raw chicken meat isolates (Figure 3).
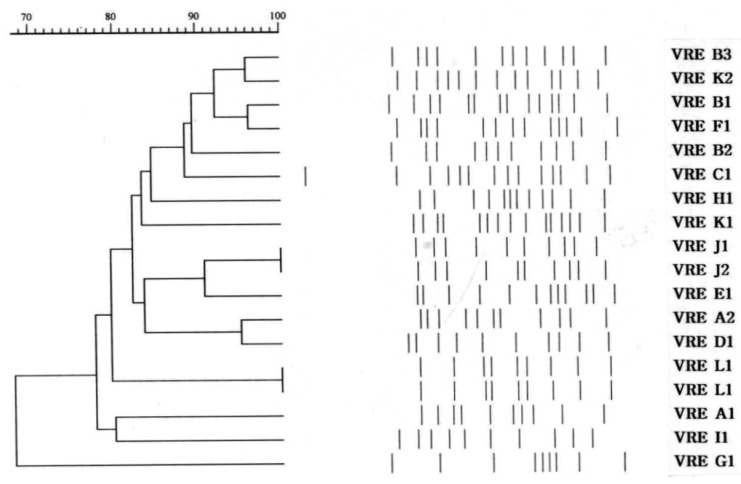
On dendrogram analysis, clinical isolates were diverse, but the raw chicken meat isolates showed relatively high similarity (Figure 4). There were no characteristic band patterns that differentiated the clinical isolates from the raw chicken meat isolates.
As a result of PFGE of vanC1 and vanC2 isolates, there were no identical bacterial strains observed and they appeared variously.
Enterococcus spp. infection including VRE has been known mainly as a hospital-acquired infection occurring in groups through sporadic infection or the transmission of clonal bacterial strains. Different from the US, there have been reports in many European countries, such as England, Belgium, and the Netherlands, that although VRE isolation rate is low, VRE is being colonized in the intestinal tract through livestock or pets in the community4, 11, 16). Based on the fact that most of the nosocomial VRE bacterial strains have different genetic heterogeneities from one another, it was first thought in Europe that VRE might have entered from the community, and it has been verified by many reports that genetically associated VRE bacterial strains have been isolated from livestock on farms, raw meats, and healthy individuals9, 10). As stated in the introduction, avoparcin, a glycopeptide antibiotic, has been added in the feeds of livestock to be used as growth promoter in Europe4). Thus, VRE is speculated to have spread through the livestock. In the US, on the other hand, many studies are reporting that there is a very small possibility that VRE had spread from healthy individuals in the community, and that avoparcin has never been used as a growth promoter for the livestock9). Also, the amount of vancomycin used in the US is 5 times as much as that of the European countries, and since oral vancomycin use is not being restricted, the glycopeptide resistance rate is considered high in the clinical field10). Actually, there has been a report of a clinical study by Auwera et al. that administered oral vancomycin into a subject, and as a result, vanA VRE was selectively colonized in the feces11). Domestically, VRE has been considered to cause nosocomial infections as it had in America since vancomycin-resistant E. durans was first reported in a patient with acute leukemia in 1992. However, avoparcin had been used in Korea from 1983 to 1998, and since Park et al. have reported 2.58% of VRE colonization rate (10 vanA, 1 VanB) in the study with 32 chickens and 70 pigs6), spreading of VRE among healthy individuals in the community through the livestock cannot be excluded. VRE can easily spread, as it can survive for about 7 days in the environment, and its carriers, while carrying VRE for 2 years, can spread it through the feces. Therefore, dissemination of VRE in the community is said to have a very important clinical significance12). As reported in this study and many others13, 15), VRE was cultivated in the suspension culture of BHI broth and it was then cultivated again in the selective medium (enterococcosel agar) in order to increase its detection rate. In Europe, however, the subsisting rate of VRE in the healthy individuals in the community is being reported from up to 5% to 12~28%4, 18) whereas this study showed as low as 1%, compared to 14% in the livestock and 60% in the raw chicken meat.
Although the identification of bacterial strains was not mentioned in the results, the phenotype identification method that used API 20 STREP (bioMerieux, Marcyl' Etoile, France) and the genotype identification method showed differences in the results of the livestock. In the identification that used API 20 STREP, most of the cases that were identified as E. faecium were verified as E. gallinarum or E. casseliflavus as a result of PCR. This seems to be due to the differences caused by the glucose fermentation between the human body isolates and livestock isolates, as presented in the study by Beatriz et al.14). As a result of the bacterial strain identification, as known from before, E. gallinarum and E. casseliflavus have rated high compared to E. faecium or E. faecalis in the livestock, but all were vanA type E. faecium in the clinical isolates.
The most important characteristics of Enterococcus spp. are not only that it essentially shows its intrinsic resistance to various kinds of antibiotics, but also that it easily acquires resistance. Enterococcus spp. produces PBP (penicillin-binding protein), which has low binding affinity to the ╬▓-lactam antibacterial agent, and shows resistance because of the tolerance (the trait that the bactericidal effects are not shown even if the growth has been suppressed once) one exposure to the ╬▓-lactam antibacterial agent, and therefore, use of the ╬▓-lactam antibacterial agent alone is difficult to expect. Additionally, in this study, it was shown that resistance to the ╬▓-lactam antibacterial agent was not due to ╬▓-lactamase production by showing a low ╬▓-lactamase production rate. Also, aminoglycoside mostly shows a low degree of resistance due to the inactivating enzyme (6-acetyltransferase) produced by Enterococcus spp. and the low permeability to Enterococcus spp. Therefore, only gentamicin and streptomycin can be used therapeutically in many cases, and it shows a high degree of resistance in some cases, due to the plasmid-mediated gene transfer and ribosomal mutation. Vancomycin shows disinfecting effects by blocking the transglycosylation and transpeptidation steps that are needed for the multimerization of peptidoglycan by binding with D-alanyl-D-alanine, the peptidoglycan precursor in the process of cell wall synthesis of gram-positive cocci, and resistance is induced by the ligase activity which shows modified matrix-specificity25). As such, VRE mostly shows resistance to conventional antibiotics. Thus, there is no effective therapeutic agent, and it is perceived to be a very serious pathogenic bacterium because vancomycin-resistant Staphylococcus aureus (VRSA) can emerge through the transferring of vancomycin-resistant genes to S. aureus. In Japan, VRSA with higher than 8 ┬Ąg/mL of MIC to vancomycin was actually isolated from a patient for the first time in 199616). Afterwards, it was also found in the US, France, and domestically, and this issue is being raised as a serious problem. In this study, according to the resistant genotype test, all of the nosocomial isolates and raw chicken meat isolates showed resistance to most of the antibiotics with vanA genotype, but healthy individual isolates and the clinical isolates obtained from patients mostly showed susceptibility to tetracycline. Almost all of the isolates showed susceptibility to chloramphenicol. Such findings are identical to the report by Choi et al18). The causes of these differences are thought to be that tetracycline-type antibiotics are still frequently being used as growth promoters for livestock, but its use has been decreased in humans. It is also considered to be due to side effects, such as aplastic anemia in the case of chloramphenicol, although it is rare, and its use has been restricted. Therefore, tetracycline and chloramphenicol can be considered as treating agents. According to the result of the testing of the high resistance of vanA-type VRE to aminoglycoside antibiotics, the livestock and raw chicken isolates showed high resistance rates to streptomycin when compared to the human isolates, while showing low resistance rates to gentamicin. It is believed that this is because streptomycin is being used for treatment purposes in livestock, but gentamicin is not being used. The results from the phenotyping of vanA, vanB, and vanC types using MIC and the results from the genotyping of vanA, vanB, and vanC types using PCR were identical. In this study, mostly vanC-type VRE was isolated in the cases that used the fecal samples of the livestock, while vanA-type E. faecium was isolated in the cases of the raw chicken meat samples that used intestinal secretion as the specimen. Based on this, the possibility of the selective spread and isolation of highly resistant vanA VRE in the manufacturing process of raw chicken meat or differences in the sample-collecting process can be considered.
As a result of analyzing the patterns of PFGE of vanA VRE, clinical isolates were variously manifested, and there was no particular type that appeared more frequently. However, 9 isolates from the raw chicken meat samples showed findings that were identical in 3 PFGE patterns, and 1 bacterial strain showed identical PFGE patterns with the livestock isolates. Therefore, in addition to the vertical transmission of the same clone, the possibility of both vertical transmission of resistant bacterial strains and horizontal transmission of resistant genes can be also considered. Actually, in the beginning, vertical transmission was considered the main mechanism for vancomycinresistant transmission. However, although epidemiologically related, due to the many cases that show various PFGE patterns, resistance by the horizontal transmission of vanA genes is now also known to be a main mechanism18, 29, 30). To investigate the epidemiology of VRE, further study is needed. These studies must include vanA gene cluster analysis (overlapping PCR), southern hybridization analysis, and conjugation study of gene transfer.
Considering the remarkably low VRE isolation rate in healthy individuals compared to the high isolation rate of vanA resistant genotype in the raw chicken meat and the differences in the results of susceptibility of the animal and human body isolates, the possibility for the vertical transmission of resistant bacteria or the horizontal transmission of resistant genes from animals to humans is very low. Considering the similar PFGE patterns of the raw chicken isolates, VRE contamination during the manufacturing process of raw chicken meat can be considered. It is possible that VRE cross-contamination may occur through poor handwashing or improper cleansing of butchers' kitchenboards and knives. The fecal specimen was taken by swabbing of stool samples, but intestinal secretion of chicken meat was taken by rinsing after managing in the microwave oven for 1 minute. The differences in the isolation rates might be influenced by the specimen-collection method. Domestically, however, there is a mixed epidemiological trait between the European trait, where antibiotics such as avoparcin and tylosin were used as growth promoters for livestock, and the American trait, where the antibiotics are being widely used in hospitals. Also, the isolation rate might have been low because the bacterial cultivation was done with fecal sample cultures instead of intestinal secretions. Therefore, larger scale studies should be conducted in the future.
Notes
This study was supported by a grant from the Korean Society of Internal Medicine, and Technology Development Program for Agriculture and Forestry(201102-03-2-HD120), Ministry of Agriculture and Forestry, Republic of Korea.
References
1. Murray BE. The life and times of the enterococcus. Clin Microbiol Rev 1990;3:46ŌĆō65PMID : 2404568.



2. Schaberg DR, Culver DH, Gaynes RP. Major trends in the microbial etiology of nosocomial infection. Am J Med 1991;91(3B):72SŌĆō75SPMID : 1928195.

3. Leclercq R, Derlot E, Duval J, Courvalin P. Plasmid-mediated resistance to vancomycin and teicoplanin in enterococcus faecium. N Engl J Med 1988;319:157ŌĆō161PMID : 2968517.


4. Goossens H. The epidemiology of vancomycin-resistant enterococci. Curr Opin Infect Dis 1999;12:537ŌĆō541PMID : 17035818.


5. Kim BS. Vancomycin-resistant enterococci in the regional society. CDMR 2002;13:37ŌĆō41.
6. Park YH, Seo KS, Yoo HS. Development of multiplex PCR for detection of vancomycin-resistant enterococci and epidemiological application in Korea. J Korean Soc Chemother 1999;17:369ŌĆō384.
7. NCCLS. Performance standards for antimicrobial susceptibility testing: 9th international supplement NCCLS document 19:M100-S109. 1999.
8. Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol 1995;33:2233ŌĆō2239PMID : 7494007.



9. Kim WJ. Global epidemiology of glycopeptide-resistant enterococci: an update. Proceedings of 3rd International Symposium on Antimicrobial Agents and Resistance. 2001;Seoul: 129ŌĆō139.
10. Sundsfjord A, Simonsen GS, Courvalin P. Human infections caused by glycopeptide-resistant enterococcus spp.: are they a zoonosis? Clin Microbiol Infect 2001;7:16ŌĆō33PMID : 11688531.

11. van der Auwera P, Pensart N, Korten V, Murray BE, Leclercq R. Influence of oral glycopeptides on the fecal flora of human volunteers: selection of highly glycopeptide-resistant enterococci. J Infect Dis 1996;173:1129ŌĆō1136PMID : 8627064.


12. Shin DH, Park JH, Shin JH, Suh SP, Ryang DW, Kim SJ. A molecular epidemiologic study on vancomycin-resistant enterococci isolated from clinical specimens over a 5-year period. Korean J Infect Dis 2000;32:203ŌĆō211.
13. Ieven M, Vercauteren E, Descheemaeker P, van Laer F, Goosens H. Comparison of direct plating and broth enrichment culture for the detection of intestinal colonization by glycopeptide-resistant enterococci among hospitalized patients. J Clin Microbiol 1999;37:1436ŌĆō1440PMID : 10203501.



14. Robredo B, Singh KV, Baquero F, Murray BE, Torres C. Vancomycinresistant enterococci isolated from animals and food. Int J Food Microbiol 2000;54:197ŌĆō204PMID : 10777070.


15. D'Agata EM, Jirjis J, Gouldin C, Tang YW. Community dissemination of vancomycin-resistant Enterococcus faecium. Am J Infect Control 2001;29:316ŌĆō320PMID : 11584258.


16. Kanchana MV, Dencer H, Blondeau J. Cost-effective algorithm for detection and identification of vancomycin-resistant enterococci in surveillance cultures. Eur J Clin Microbiol Infect Dis 2000;19:366ŌĆō369PMID : 10898139.


17. Wootton M, Howe RA, Hillman R, Walsh TR, Bennett PM, MacGowan AP. A modified population analysis profile (PAP) method to detect hetero-resistance to vancomycin in Staphylococcus aureus in a UK hospital. J Antimicrob Chemother 2001;47:399ŌĆō403PMID : 11266410.


18. Choi YH, Lee YS, Lee JK, Yoo JL, Kim CK, Kim BA. Molecular relationship of vanA glycopeptide resistance gene in enterococci from hospitalized patients and poultry in Korea. Korean J Infect Dis 2001;33:383ŌĆō391.
19. van den Bogaard AE, Jensen LB, Stobberingh EE. Vancomycinresistant enterococci in turkey and farmers. N Engl J Med 1997;337:1558ŌĆō1559PMID : 9380129.


20. Cheong HJ, Kim WJ, Woo HJ, Kim MJ, Park SC. Study on the infection due to VanA type vancomycin-resistant enterococci. Korean J Infect Dis 1998;30:10ŌĆō18.
21. Kim DR, Yu CW, Cheong HJ, Woo HJ, Kim WJ, Kim MJ, Park SC, Choi SJ. Rectal surveillance culture of vancomycin-resistant enterococci colonization among patients hospitalized in the intensive care unit. Korean J Infect Dis 1999;31:203ŌĆō209.
22. Cheong HJ, Kim WJ, Choi SJ, Lee KW, Choi KW, Park SC. Nationwide questionnaire survey for the prevalence, detection methods, and infection control program of VRE among hospitals. Korean J Infect Dis 2001;33:78ŌĆō87.
23. Arthur M, Courvalin P. Genetics and mechanisms of glycopeptide resistance in enterococci. Antimicrob Agents Chemother 1993;37:1563ŌĆō1571PMID : 8215264.



24. van den Bogaard AE, Willems R, London N, Top J, Stobberingh EE. Antibiotic resistance of faecal enterococci in poultry, poultry farmers and poultry slaughterers. J Antimicrob Chemother 2002;49:497ŌĆō505PMID : 11864950.


25. van den Bogaard AE. relation to human and animal exposure to antibiotics. J Antimicrob Chemother 1997;40:453ŌĆō454PMID : 9338504.


26. Murray BE. Vancomycin-resistant enterococcal infection. N Engl J Med 2000;342:710ŌĆō721PMID : 10706902.


27. Park JW, Kim YR, Shin WS, Kang MW, Han KJ, Shim SI. Susceptibi1ity tests of vancomycin-resistant enterococci. Korean J Infect Dis 1992;24:133ŌĆō137.
28. Edmond MB, Ober JF, Dawson JD, Weinbaum DL, Wenzel RP. Vancomycin-resistant enterococcal bacteremia: natural history and attributable mortality. Clin Infect Dis 1996;23:1234ŌĆō1239PMID : 8953064.


Figure┬Ā2
Comparison of antibiotic resistance rates of vanA E. faecium isolates between people and animals. (TE: tetracycline, EM: erythromycin, AM: ampicillin, CM: chloramphenicol, GM: gentamicin, SM: streptomycin)
Figure┬Ā3
PFGE patterns of vanA E. faecium isolates (A1~I1: human, J1~K2: raw chicken meat, L1: animal and raw chicken meat).
Figure┬Ā4
Dendrogram of vanA E. faecium isolates (A1~I1: human, J1~K2: raw chicken meat, L1: animal and raw chicken meat).
Table┬Ā1
Primer sequences for PCR of vancomycin resistance genotyping and enterococci species identification among vancomycin-resistant
enterococci.




 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Download Citation
Download Citation Print
Print



