 |
 |
| Korean J Intern Med > Volume 8(2); 1993 > Article |
|
Abstract
The authors investigated the distribution of HCV genotypes in patients with various chronic liver diseases in Korea. Study population was 70 individulals, positive for second generation anti-HCV EIA, consisting of 37 cases with sporadic non-A, non-B (NANB) chronic hepatitis (CH), 12 NANB hepatocellular carcinoma, 16 post-transfusion NANB hepatitis, 4 non-B blood donors and 1 healthy family member of a patient with sporadic CH. Molecular typing was performed by RT-nested PCR with type-specific primer sets deduced from the NS-5 region of HCV. The prevalence of type II was 75.0% and type III was 25.0% in sera. In liver tissues, type II HCV was shown in 63.0%, type III HCV in 3.7% and co-infections with type II and III HCV were observed in 18.5% of 27 samples biopsied. In the sera of patients with chronic hepatitis, typing results were relatively well correlated with those in tissues (75%), but type III could not be observed. Among 12 HCC patients, type III HCV appeared only in tissues, not in sera. These results suggest that type II HCV may be the major HCV type in Korea, and co-infections with type II and-III HCV may not be rare in chronic liver diseases with HCV.
Since the hepatitis C virus (HCV) has been isolated by Choo et al. (1989)1) and the immunoassay for HCV using antigen expressed by a region of HCV cDNA has been developed2), HCV is recognized as the major causative agent of non-A, non-B (NANB) hepatitis. Recently, many HCV strains have been isolated by sequencing completely or partially from different area of the world. The complete genomic sequences are now available for at least four HCV isolates3ŌĆō6), and several kinds of HCV genotype on the basis of partial sequencing of HCV genome are reported7ŌĆō11). By these results, HCV types could be classified into at least 4 or more types on the basis of sequence variations6ŌĆō12) and also 2 groups on the basis of group-specific antigenicity in HCV polypeptides of the NS3-4 regions have been proposed113).
As several kinds of HCV typing methods are being developed7,13ŌĆō16), the differences in the geographical distribuion of each type were observed and the possibility that individual types may be different in both the viral pathogenecity and the clinical courses has been also suggested17,18).
In this study, to investigate the distribution of HCV genotypes in patients with various chronic liver diseases positive for anti-HCV in Korea, we applied a typing of the HCV method using reverse transcription-nested polymerase chain reaction (RT-nested PCR) with type-specific primer sets deduced from the NS5 region of HCV gemome14) and we present the results of the HCV genotyping.
Sera or liver tissues were obtained from 70 patients with chronic liver diseases who consisted of 37 sporadic chronic hepatitis (CH), 12 hepatocellular carcinoma (HCC), 16 post-transfusion NANB hepatitis (PT-NANBH), 4 blood donors negative for HBsAg and 1 healthy family member of a patient with sporadic CH. All patients showed positivity in the second generation anti-HCV enzyme immunoassay (Anti-HCV-II EIA; Abbott Laboratories, Chicago, IL., U.S.A.). Frozen liver specimens, biopsied, were available in 8 sporadic CH and 12 HCC.
RNA from sera or liver tissues was extracted by acid-guanidinium-phenol method as described previously19). Briefly, 200╬╝l of serum was mixed with 200 ul of solution-D (4 M guanidinium thiocyanate, 100 mM Tris-HCl, pH 7.5; 0.5% N-lauroylsarcosine; 0.1 M beta-mercaptoethanol) and 2 M sodium acetate (pH 4.5). A piece of frozen liver tissue biopsied was homogenized in 400 ul of solution-D by DounceŌĆÖs homogenizer, and 2 M sodium acetate (pH 4.5) was added. After phenol/chloroform extraction and then isopropanol precipitation, the pellet was dissolved in 20 ╬╝l of 1% DEPC-treated distilled water.
An aliquot (4 ul) of RNA solution was mixed with 10 ╬╝l mixture containing 10 pmole of the outer antisense primer (#167 R, Table 1), 50 mM tris-HCl (pH 7.5), 75 mM KCl, 3 mM MgCl2, 0.5 mM of each dNTP, then heat treated for 5 min at 70┬░C. 20 units of Moloney Murine Leukemia Virus Reverse Transcriptase (MMLV-RT; BRL, Gaithersburg, Md, U.S.A.) was added and incubated at the mixture for 60 min at 37┬░C, then 10 min at 90┬░C
Oligonucleotides were synthesized by DNA synthesizer (Pharmacia LKB Gene Assembler Plus, Uppsala, Sweden).
Oligonucleotide primers deduced from the NS5 region of the HCV genome were five sets, including an outer or universal primer set (401 bp) and four type-specific primer sets. The sequences of primers used as outer set in the first round PCR was #166 and #167R, which correspond to positions nucleotide (nt) 8230 to 8260 and the complementary sequence to nt 8601 to 8630 of HCV-J, respectively3).
To determine the genotype of HCV-J, -US, -K1 and -K2 in the second round PCRs, the primers which were designed by Enomoto et al.,7) and Kato et al.,14), #192 (nt 8291 to 8310, sense) and #193R (nt 8487ŌĆō8506, antisense) for HCV-J, #194 (nt 8291 to 8310, sense) and #195R (nt 8487 to 8506, antisense) for HCV-US, #173 (nt 8281 to 8301, sense) and # 174R (nt 8462 to 8481, antisense) for HCV-K1, #151 (nt 8283 to 8302, sense) and #153R (nt 8460 to 8479, antisense) were used (Table 1). The expected sizes of PCR products for each type, HCV-US, HCV-J, HCV-K1 and HCV-K2 were 216 bp, 216 bp, 201 bp and 197 bp, respectively, and they were respectively corresponding to type I, -II, -II, and -III on the proposed phylogenic classification of HCV7,12,14)(Fig. 1).
In the first PCR, 50 ╬╝l mixture, containing 10 mM tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 0.01% gelatin, 2.5 units of recombinant Taq. DNA polymerase (Pharmacia, Biotech. U.S.A.), 200╬╝M each dNTP, 0.1 ╬╝M of each outer primer and 10 ╬╝l of cDNA was amplified by Thermocycler (Perkin-Elmer-Cetus Co., LA, U.S.A.) for 35 cycles. Each cycle included denaturation at 94┬░C for 1 min, primer annealing at 40┬░C for 1 min and primer extension at 72┬░C for 1 min. Then, with 1 ╬╝l of the first PCR products in the prepared 4-each type specific microtube, the second PCR was carried out for 30 cycles with denaturation for 1 min at 94┬░C, primer annealing for 45 seconds at 55┬░C and primer extension for 1 min at 72┬░C. The products of the second PCR were subjected to electrophoresis on 2% agarose gel (BRL, Gaithersburg, Md, U.S.A.), stained with ethidium bromide and observed under U.V. light.
37 patients with sporadic CH and 4 blood donors all showed positivity in RT-nested PCR, while 9 (56.2%) of 16 cases with post-transfusion hepatitis were positive. Among 12 patients with HCC, 5 cases (62.5%) of 8 sera, 10 cases (83.3%) of 12 frozen tumorous tissue specimens and 5 cases (71.4%) of 7 non-tumorous tissue specimens were positive. A healthy person who was positive for anti-HCV and normal in ALT level showed negativity in RT-nested PCR.
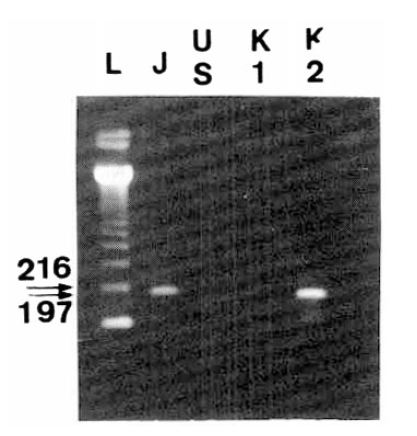
The strategy for gonotyping HCV is illustrated in Fig. 2. Type specific PCR products, obtained through the procedure of RT-nested PCR, were visualized after they were electrophoresed and stained with ethidium bromide (Fig. 3).
Among a total 70 individuals, available 43 serum samples were typed by RT-nested PCR. The prevalence of type II HCV was 75%, type III HCV was 25% and there was no co-infection with type II and -III HCV. 24 cases (64.8%) of 37 sporadic CH were type II HCV, while 13 cases (35.2%) were type III HCV.
All cases of 6 HCC and 9 PT-NANBH showed only type II HCV and 3 cases (75.0%) of 4 blood donors were type II HCV, while 1 case (25.0%) was type III HCV (Table 2).
In frozen liver tissues, type II HCV was 63.0%, type III HCV was 3.7% and co-infection with type II and-III HCV were shown in 18.5%(Table 3, Fig. 4).
Among 27 cases which were obtained by liver biopsy, 6(75.0%) of 8 sporadic CH, 7(58.3%) of 12 tumorous tissues and 4(57.1%) of 7 non-tumorous tissues of HCC. Co-infection with type II and -III HCV was 25.0% in sporadic CH, 16.7% in tumorous tissues of HCC and 14.3% in non-tumorous tissues of HCC, respectively.
8 liver tissue specimens were obtained in sporadic chronic patients. In sera, all 8 cases showed type II HCV, while in tissues, 6 cases were type II and 2 cases were co-infection with type II and -III HCV. Typing results in sera were relatively well correlated with those in tissues (75%), but the positivity in type III could not be observed (Table 4).
In sera, 6 cases which were positive in RT-nested PCR among 12 HCC patients, were all type II HCV, while, in tuomorus liver tissues, type II HCV was 7 cases, type III HCV was 1 case, co-infections with type II and -III HCV were 2 cases and, in non-tumorous tissues, type II was 4 cases and co-infections were 1 case (Table 5).
HCV, which has been known as a major causative agent of NANB hepatitis and is responsible for development into cirrhosis and HCC1,20), contains a positive-stranded RNA genome of about 9,400 nucleotides1) and a single, large translational open reading frame (ORF) which encodes a viral polypeptide of 3,011 amino acids4).
Recently, as several kinds of HCV typing methods are being developed7,13ŌĆō16), many HCV stains have been isolated from different areas of the world, and the differences in the geographical distribution of each type have been observed, and the possibility that individual types may have differences in both the viral pathogenecity and the clinical courses, have been also suggested17,18).
Among the many HCV isolates in other countries, type I HCV has been considered to be a major type in the United states and type II HCV is reported as the most prevalent type in Japan3,4,6,14). Also, In European countries, the HCV isolates from Germany and France were closely related to the prototype HCV from United States (type 1 or HCV-US)22,23). Pozzato et al. revealed at least two HCV types in Italy, type II HCV and other unknown type24), and in Thailand12) HCV-V and -IV have been isolated. But, korea, systemic and epidemiological studies for HCV type were not fully performed until now, even if some report revealed a full sequence of HCV genome in Korean patients21).
In an effort to identify types of HCV isolated in Korean patients, we performed typing of HCV in sera and liver tissues by using the RT-nested PCR with type-specific primers.
In this study, we found that the most pevalent HCV type in Korea was type II like Japan, in which type II were 70% to 80%14,15). According to the authors, the homologies of type II HCV isolated from Korean patients to representative reference sequences12,14,15) isolated from Japan were relatively high, more than 90% at the nucleic acid level and 95% at the amino-acid level (unpublished). This suggested that HCV strains in both countries may have close relationship in the phylogenetic evolution of HCV, and further studies are required to clarify that issue.
It has been reported that the prevalence of type III HCV in Japan was 10% to 26%17,25) and cases with co-infection with II and -III were rare25). In our study, we couldnŌĆÖt find any case showing co-infection with type II and -III in serum, but 25% of the cases with available liver tissues biopsied showed co-infection. These results suggested that the relatively high proportion of the cases may be infected by two or more HCV strains. In the correlation of HCV types between sera and tissues, type II HCV appeared predominantly in both, but type III was presented only in liver tissue. The reason why is not clear but there may be the possibility that the replicative activity of type III HCV may be lower than type II. The clinical significance according to HCV types has not been studied fully, but some reports revealed the association between HCV types and clinical features17,18,25). Takada et al.17) reported that type III HCV might be more frequently infected in sporadic forms than type II and the inital response to interferon treatment was better in patients with type III HCV than those with type II HCV. From these results they suggested the possibility that the effects of interferon might be different according to the types of HCV. In our previous data, we found that there was a discrepancy on effects of interferon treatment even in the same type II HCV and that sporadic chronic patients with type II HCV could be divided into two groups by the responsiveness to recombinant interferon-alfa administration, responder and non-resonder group. The non-responder group tended to show more severe clinical courses than the responder group. Therefore it suggests that HCV types would be useful as one of prognositc factors determining response to interferon treatment. However, further study for the clinical implication of HCV types have to be evaluated in a larger number of patients and in various geographical areas where different HCV types are isolated.
Fig.┬Ā2.
Schematic diagram for typing HCV by PCR with type specific primers. Spotted blocks indicate different productŌĆÖs size that were specific to each of the four HCV types.
Fig.┬Ā3.
Second PCR products for typing HCV. Sequences in the NS5 gene of different HCV types were amplified by RT-nested PCR with type specific primers, subjected to elcetrophoresis and stained with ethidium bromide. (A) and (B) showed HCV-J and HCV-J/K1, respectively. These corresponded to type II HCV. (C) indicated HCV-K2, corresponding to type III HCV. The letter ŌĆ£LŌĆØ indicated molecular size markers (123 DNA ladder, BRL, U.S.A).

Fig.┬Ā4.
Coinfections of HCV in liver tissue from a patient with chronic hepatitis. Two positive bands on HCV-J and HCV-K2 indicated a mixed infection with type II and type III. The letter ŌĆ£LŌĆØ indicated molecular size marker (123 DNA ladder; BRL, U.S.A)
Table┬Ā1.
Oligonucleotide Primers Deduced from a Part of NS5 Region of HCV Genome
Table┬Ā2.
Molecular Typing Analysis of HCV Genome in Sera-Using RT-Nested PCR with Type-Specific Primers
Table┬Ā3.
Molecular Typing Analysis of HCV Genome in Frozen Liver Tissue Specimens Using RT-Nested PCR with Type-Specific Primers
Table┬Ā4.
Correlation of HCV Types Between Serum and Tissue is Each Chronic Hepatitis Patient
Table┬Ā5.
Correlation of HCV Types Between Serum and Tissue in Each Hepatocellular Carinoma Patient
| No | Age/Sex | Serum | Tumor Site | Nontumor Site |
|---|---|---|---|---|
| #1 | 60/M | (ŌłÆ) | type II | n.t.* |
| #2 | 77/M | (ŌłÆ) | type II | n.t.* |
| #3 | 72/M | type II | type II+III | type II |
| #4 | 58/M | type II | type II | n.t.* |
| #5 | 67/M | n.t.* | type III | (ŌłÆ) |
| #6 | 54/M | n.t.* | (ŌłÆ) | type II |
| #7 | 66/M | n.t.* | type II | (ŌłÆ) |
| #8 | 46/M | type II | (ŌłÆ) | type II |
| #9 | 54/M | n.t.* | type II | n.t.* |
| #10 | 47/M | type II | type II | type II |
| #11 | 66/M | type II | type II | type II+III |
| #12 | 69/M | type II | type II+III | n.t.* |
REFERENCES
1. Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M. Isolation of cDNA clone derived from a blood borne non-A, non-B viral hepatitis genome. Science 244:359. 1989.


2. Kuo G, Choo QL, Alter HJ, Gitnick GL, Redeker AG, Purcell RH, Miyamura T, Dienstag JL, Alter MJ, Stevens CE, Tegtmeier GE, Bonino F, Colombo M, Lee WS, Kuo C, Berger K, Shuster JR, Overby LR, Bradley DW, Houghton M. An assay for circulating antibodies to a major etiologic virus of human non-A, non-B hepatitis. Science 244:362. 1989.


3. Kato N, Hijikata M, Ootsuyama Y, Nakagawa M, Ohkoshi S, Sugimura T, Shimotohno K. Molecular cloning of the human hepatitis C virus genome from Japanese patients with non-A, non-B hepatitis. Proc Natl Acad Sci USA 87:9524. 1990.



4. Choo QL, Richman KH, Han JH, Berger K, Lee C, Dong C, Gallegos , Coit D, Medina-Selby A, Barr PJ, Weiner AJ, Bradley DW, Kuo G, Houghton M. Genetic organization and diversity of the hepatitis C virus. Proc Natl Acad Sci USA 88:2451. 1991.



5. Takamizawa A, Mori C, Fuke I, Manabe S, Murakami S, Fujita J, Onishi E, Andoh T, Yoshida I, Okayama H. Structure and organization of the hepatitis C virus genome isolated from human carriers. J Virol 65:1105. 1991.



6. Okamoto H, Okada S, Sugiyama Y, Kurai K, Lizuka H, Machida A, Mayumi M. Nucleotide sequence of the genomic RNA of hepatitis C virus isolated from a human carrier: Comparison with reported isolates for conserved and divergent regions. J Gen Virol 72:2697. 1991.


7. Enomoto H, Okada S, Sugiyama Y, Data T. There are two major types of hepatitis C virus in Japan. Biochem Biophys Res Commun 170:1021. 1990.


8. Okamoto H, Okada S, Sugiyama Y, Yotsumoto S, Tanaka T, Yoshizawa H, Tsuda F, Miyakawa Y, Mayumi M. The 5-terminal sequence of the hepatitis C virus genome. Jpn J Exp Med 60:167. 1990.

9. Kubo Y, Takeuchi K, Boonmar S, Katayama T, Choo QL, Kuo G, Miyamura T. A cDNA fragment of hepatitis C virus isolated from an implicated donor of post-transfusion non-A, non-B hepatitis in Japan. Nucleic Acids Research 17:10367. 1989.



10. Takeuchi K, Kubo Y, Boonmar S, Watanabe Y, Katayama T, Choo QL, Kuo G, Floughton M, Saito I, Miyamura T. The putative nucleocapsid and envelope protein genes of hepatitis C virus determined by comparison of the nucleotide sequences of two isolates derived from an experimentally infected chimpanzee and healthy human carriers. J Gen Virol 71:3027. 1990.


11. Ogata N, Alter HJ, Miller RH, Purcell RH. Nucleotide sequence and mutation rate of the H strain of hepatitis C virus. Proc Natl Acad Sci USA 88:3392. 1991.



12. Mori S, Kato N, Yagyu A, Tanaka T, Ikeda Y, Petchclai B, Chiewsilp P, Kurimura T, Shimotohno K. A new type of hepatitis C virus in patients in thailand. Biochem Biophy Res Commun 183:334. 1992.

13. Tsukiyama-Kohara K, Yamaguchi K, Maki N, Ohta Y, Miki K, Mizokami M, Ohba KI, Tanaka S, Hattori N, Nomoto A, Kohara H. Antigenecities of group I and II hepatitis C virus polypeptides-Molecular basis of diagnosis. Virology 192:430. 1993.


14. Kato N, Ootsuyama Y, Ohkoshi S, Nakazawa T, Mori S, Hijikata M, Shimotohno K. Distribution of plural HCV types in Japan. Biochem Biophys Res Commun 181:279. 1991.


15. Nakao T, Enomoto N, Takada N, Takada A, Date T. Typing of hepatitis C virus genomes by restriction fragment length polymorphism. J Gen Virol 72:2105. 1991.


16. Okamoto H, Kurai K, Okada S, Yamamoto K, Lizuka H, Tanaka T, Fukuda S, Tsuda F, Mishiro S. Full-length sequence of a hepatitis-C virus genome having poor homology to reported isolates: Comparative study of four distinct genotypes. Virology 188:331. 1992.


17. Takada N, Takase s, enomoto N, Takada A, Date T. Clinical backgrounds of the patients having different types of hepatitis C virus genomes. J Hepatol 14:35. 1992.


18. Di Bisceglie AM, Order SE, Klein JL, Waggoner JG, Sjogren MH, Kuo G, Houghton M, Choo QL, Hoofnagle JH. The role of chronic viral hepatitis in hepatocellular carcinoma in the United States. Am J Gastroenterol 86:335. 1992.
19. Kato N, Ohkoshi S, Shimotono K. Japanese isolates of non-A, non-B hepatitis viral genome show the sequence variation from the original isolate in the USA. Proc Jpn Acad 65:219. 1989.

20. Alter HJ, Purcell RH, Shih JW, Melpolder JC, Houghton M, Choo L, Kuo G. Detection of antibody to hepatitis C virus in prospectively followed transfusion recipients with acute and chronic non-A, non-B hepatitis. N Engl J Med 321:1494. 1989.


21. Cho YG, Yoon JW, Jang KL, Kim CM, Sung YC. Full genome cloning and nucleotide sequene analysis of hepatitis C virus isolates. Gene 105:167. 1991.


22. Fuchs K, Motz M, Shreier E, Zachoval R, Deinhardt F, Roggendorf M. Characterization of nucleotide sequences from European hepaptitis C virus isolates. Gene 105:167. 1991.


23. Li JS, Tong SP, Vitvitski L, Lepot D, Trepo C. Two French genotypes of hepatitis C virus: homology of the predominant genotype with the prototype American strain. Gene 105:167. 1991.






 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Download Citation
Download Citation Print
Print



