INTRODUCTION
Diabetes mellitus (DM) is a heterogeneous group of diseases that shares hyperglycemia.
Diabetes mellitus occurs due to the absolute and relative defect of insulin action, and disease pathogenesis is partially suggested that disease expression is closely correlated with modulation of its gene action or other gene abnormality.
DM is usually classified as two types.1) These are insulin dependant DM (IDDM, type I) and non-insulin dependant DM (NIDDM, type II). Notably IDDM is caused by viral insulitis, specifically correlated with HLA gene, islet cytoplasmic antibody (ICA), and we thought that IDDM might have been an autoimmune disease because islet cell surface antibody (ICSA) is detected in the early IDDM patientŌĆÖs serum.
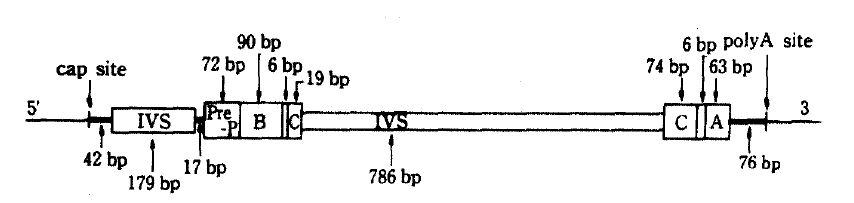
NIDDM has many of the features of a genetic back ground, some report had proved confidently in the study of identical twin and found 100 percent identical rates of the disease.2) But the precise mechanism of the NIDDM genetic defects are not known, the amino-acid sequence of human insulin has been known since 1959,3) but it was not until 1980 that the nucleotide sequence of the human insulin gene was determined.4) Nowadays insulin gene region of two homologues is located band p15 of the short arm of human chromosome 11 has been found and charaterized.6) Also highly polymorphic region if found 5ŌĆ▓-flanking region near the initiation of transcription of insulin mRNA begins, and RFLP has been described in Caucacians,7ŌĆō12) American Blacks,13) Pima Indians14) and some other racial groups. At that region, RFLP of insulin gene is generated by the insertion-deletion of DNA sequences, so that fragments of different sizes are generated by digestion with appropriate endonucleases (for example, Pvu II, Rsa I). The interesting structure of the RFLP region is a tandemly repeated 14 base pairs ACAGGGGTGTGGGG on the upstream of transcription unit.15) On the use of endonuclease, Pvu II, cleavage sites are recognized on the polymorphic region, not including insulin structrural gene. The structure of insulin gene (Figure 1) has 1,430 base pairs (bp) and consists of a mRNA precursor that arise from a cap site and terminates at poly A adenylation sites. Also its structural gene consists of 3 exons and 2 intervening sequences (IVS) of 179 and 786 nucleotide respectvely.4) After excision of the two IVS, the mRNA molecule for preproinsulin is formed. The structure of the insulin gene flanking sequences has also been determined about 6,000 bp down stream (3ŌĆ▓) and 15,000 bp upstream (5ŌĆ▓) to the insulin gene are regions consisting of repetitive sequences, members of so called Alu-family.11ŌĆō15) The Alu family members are 300 bp repititions, which comprise 3 percent of the total human DNA,16) distributed in the whole genome. The function of Alu-family is unknown, but these seqences are considered to flank active region of chromatin.17) The Alu-family sequence and the 3ŌĆ▓-reigion are non-polymorphic,11,15) where as the 5ŌĆ▓-regin is highly polymorphic,11) i.e. variable in length. The polymorphic 5ŌĆ▓-region starts only 363 bp upstream to the insulin gene. RFLP would be found at 0.6 (class 1), 1.3 (class 2) and 2.4 kb (class 3) region respectively.11) Class 3 allele was appeared relating to disease entity, atherosclerosis or hyperlipidemia in the NIDDM patients,18) and a possible association of the gene allele (class 3) and NIDDM has also been reported; other subsequent studies have suggested that there is no association between the large allele and NIDDM in various races.11ŌĆō14,18) There are many features of racial differences in the distribution of the size classes of the restriction fragments. Class 2 fragments occur in <1% of Caucasoids, in contrast to 22% of Blacks, and class 3 fragments are frequent in Caucasoids, but rare in Asians.10,15) Class 3 allele is close related to high level of glycosylated hemoglobin (HbA1c), but exact role of this allele is not known yet. Especially in the view point of geographical resemblence between Korea and Japan, we easily guess nearly the same pattern of insulin gene RFLP which had low frequency of class 3 alleles in the Japan.19,20)
MATERIALS AND METHODS
Peripheral blood leukocytes were obtained from 40 unrelated subjects (11 nondiabetic subjects, 29 type 2 diabetic subjects). All subjects were classified according to the criteria of Natioinal Diabetes Data Group.1)
1. Preparation of DNA
Human DNA was prepared from the nuclei of white blood cells. Normally, 10 ml of blood is collected in a heparinized tube and then kept at 4┬░C. Aliquot of 5 ml are added to 50 ml disposable polyethylene centrifuge tubes and then cells are lysed by the addition of 45 ml of 0.32 M sucrose, 10 mM Tris-HCl (pH 7.5), 5 mM MgCl2 and one percent Triton X-100. The nuclei are collected by centrifgation at 1000 g for 15 min. The supernatant is discarded and each nuclear pellet is suspended in 2.25 ml of 0.075 M NaCl, 0.024 M EDTA (pH 8.0), and then combined in one of 50 ml centrifuge tube. One-halfmilliliter of a solution of 5 percent sodium dodecyl sulfate (SDS) and Proteinase K at 2 mg/ml is added and the viscous solution incubated about 12 hour at 37┬░C. The digest is then gently mixed with 5 ml of redistilled phenol saturated with 20 mM Tris-HCL (pH 8.0). Five milliliters of chloroform-isoamylalcohol (24:1) is added and vigorous mixing for 30 sec. The phases are separated by centrifugation for 15 min at 1000 g. The upper, viscous, aquous phase is after centrifugation, 0.1 volume of 3 M sodium acetate and 2 volumes of 95ŌĆō100 percent ethanol is added at room temperature. Under these conditions, DNA is selectively precipitated like thread lump and the majority of the RNA remains in the supernatsnt. Precipitated DNA placed in 1 ml of 10 mM Tris-HCl (pH 7.5), 1 mM EDTA.
2. Digestion of DNA with Restriction Enzyme
Ten micrograms of stored DNA estimated at OD260, restriction enzyme Pvu II 20 to 30 units, 10 times enzyme buffer and water are added to 100 microliter and then incubated at 37┬░C for one to two hours. The digestions are terminated by the addition of 0.1 volume of 3 M sodium acetate and DNA precipitated by adding 2 volumes of absolute ethanol. After more than 40 min in a dry ice-ethanol bath, the precipitated DNA is collected by centrifugation for 15 min, 15,000 rpm at 4┬░C. The supernatant is decanted. One millititer of 70% (v/v) chilled ethanol is added and the tubes centrifuged for 5 min as above. The superantant is decanted gently and the precipitated is dried for 10 min under reduced pressure. The dried DNA is dissolved in 15 to 20 microliter of a loading solution of 5% Ficoll 400 (Sigma), 5 mM EDTA (pH 8.0), 0.2% SDS and 0.1% bromphenol blue at room temperature. The samples are heated at 65┬░C for 10 min before being loaded on the agarose gel.
3. Agarose Gel Electrophoresis
The DNA fragments are separated according to the size in an agarose gel. We run horizontal 0.8 to 1 percent agarose gels (10 ├Ś 14 ├Ś 0.5), prepared in 0.089 M Tris-borate (pH 8.0), 0.089 M boric acid, 0.002 M EDTA at 100 volts for 5 hours. After electrophoresis, the gel is stained in 1 microgram/ml ethidium bromide for 20 min and then photographed on a 302 nanometer UV light box. The orientation of the gel should be marked by cutting a corner.
4. Transfer of DNA to A Nitorcellulose Filter
The separated DNA fragments are transfered from the agarose gel to a nitrocellulose filter paper creating replica of the pattern of DNA fragments on the filter as described by Southern.21) The DNA is denatured in situ by soaking the gel for 60 min in 1.5 M NaCl, 0.5 M NaOH. The gels are not shaken since they can fragment. The gel is then soaked in 3 M NaCl and 0.5 M Tris (pH 7.0) for 30 min and then placed on a glass plate covered with two sheets of Whatman 3 MM paper which have been soaked in 20X SSC (SSC is 0.15 M NaCl, 0.015 M sodium citrate) and then continuously soaked with same solution. The nitrocellulose filter of the same dimension as the gel are soaked in 2X SSC is laid on the gel. There should be no air bubbles between the gel and 3 MM paper of nitrocellulose filter. Two pieces of presoaked 3 MM paper and a stack of paper towels cut to the dimension of the gel are placed on the nitrocellulose filter paper. This is covered with glass plate and about 1-1b weight are placed on top. After about 24 hours, the towels are removed, and the gel is much thinner than before. The orientation can be marked on the paper with a soft pencil or ballpoint pen. The gel is separated from the nitrocellulose paper and can be restained with ethidium bromide solution to assess the amout of transferred. The nitrocellulose paper is rinsed briefly in 2X SSC, blotted, and then dried in vacuo at 80┬░C for 2 hours.
5. Labeling of the DNA probe
The 0.8 kb of eukaryotic DNA which is kindly donated from Dr. G.I. Bell can be isolated from digests of the recombinant plasmid (pBR 327 contains phins 310 in HB 101) by rapid plasmid preparaiton, preparative gel electrophoresis and electroelution in Tris-EDTA buffer, then purification of probe with chloroform-ethanol extraction as above. We labeled the DNA probe by the use of nick translation kit (Amersham). Probe DNA, nucleotide buffer, radioactive dCTP, DNAse and DNA polymerases were added and incubated at 15┬░C for 2 hours. Sephadex G-50 column chromatography was performed for the separation of 32P-bounded probe. Before chromatography, it was necessary for non-specific DNA like salmon testes DNA to be prerun to prevent adhering to the radiolabeled probe DNA. Collection of the first peaked fractions revealed bounded radioactive probe and boiled for 3 min before hybridization.
The probe can be readily labeled by nick translation procedure to specific activity 1 ├Ś 108 cpm/ug DNA.
6. Hybridization
The dry nitrocelllose filter is first presoaked in a 5X SSC and then prehybridization was performed in a solution of 5X SSC, 50 percent formamide, 50 mM sodium phosphate (pH 6.5), 0.25 percent SDS, 250 ug/ml denatured and sonicated salmon testes DNA, and 5X DenhardtŌĆÖs solution (1X DenhardtŌĆÖs solution is 0.02 percent Ficoll 400, 0.02 percent polyvinylpyrolidone 360, and 0.02 percent bovine serum albumin). After a 3 to 4 hour incubation at 50┬░C, the preannealing solution is replaced with the hybridization solution which contains 5X SSC, 50 percent formamide, 10 percent dextran sulfate, 20 mM sodium phosphate (pH 6.5), 0.25 percent SDS, 100 ug/ml denatured and sonicated salmon testes DNA, 2X DenhardtŌĆÖs solution and final concentration of 106 cpm of 32P-labeled DNA fragment probe. The specific activity of nick translated probe is usually about 1 ├Ś 107 cpm/ug DNA. The salmon testes and radiolabeled DNAs are denatured in a boiling water bath for 10 min before use. The bybridization is for 16 to 20 hour period at 50┬░C. At the end of this period, the nitrocellulose filter is removed from the hybridizaiton solution and washed in 0.1X SSC and 0.1 percent SDS at room temperature for 30 min followed by at 42┬░C for 30 min in a shaking waterbath. The nitrocellulose filter is blotted dry and the filter is wrapped with vinyl wrap and exposed to X-ray film with intensifying screen for overnight or longer at ŌłÆ70┬░C. The film is developed and investigate the RFLP of 5ŌĆ▓-flanking region of insulin gene.
RESULTS
We examined the RFLP of 5ŌĆ▓-flanking region of the insulin gene from 11 non-diabetic subjects and 29 NIDDM patients according to the criteria of National Diabetes Data Group.
Before hybridization and autoradiography, digested human DNA were run on the 0.8 ŌłÆ1 percent agaroe gels and stained with ethidium bromide (Fig. 2).
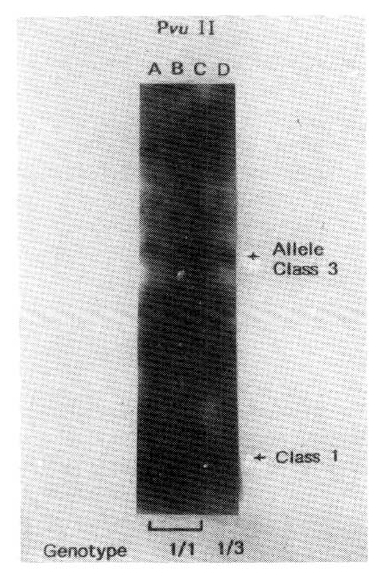
Studies of RFLP of the insulin gene region showed three different sized of DNA fragment classes of the following average size: class 1, 570 bp; class 2, 1320 bp; and class 3, 2470 bp (Fig. 3).
We have observed two major sizes of Pvu II generated RFLP in Korean; 0.6 kb and 2.4 kb. The former is considered class 1 allele, the latter is class 3 allele (Fig. 4).
Allelic frequences of the 40 subjects are shown in table 1 and large class 3 alleles had low frequency in diabetic population.
Class 3 allele and lipid profiles in 3 diabetic patients are not correlated and not any difference from other diabetics (data not shown).
We also investigated RFLP of insulin gene in four acromegalics who had secondary DM, but they revealed only class 1 allele.
DISCUSSION
Bell et al.11) noted a significant association of class 1 alleles with IDDM in Caucacians. Combined data from St. Louis, Denmark, and San Francisco confirmed this association. In this study of 27 Blacks of IDDM, the frequency of the class 1 allele was 0.70, which is not significantly different from the frequency of 0.60 in non-diabetic subjects.
In preveious studies, both Rotwein et al.10) and Owerbach et al12) found the association of class 3 alleles with NIDDM, the latter was Danish Caucacians and the former in racially mixed population. That association has not been confirmed in San Francisco. Elbein et al.22) reported no allele associated with diabetes in 313 NIDDM patients. Although class 3 homozygotes were twice as common in diabetics as non-diabetics in our study (Table 1), this difference is not statistically significant. In both studies of American Blacks and the pooled study of Caucacians, no single allele was associated with NIDDM.
Other recent studies suggested that class 3 alleles were associated with diabetic hypertriglyceridemia23) or atherosclerosis.18,23) The rationale for these studies were that insulin is one of the major hormones influencing serum triglyceride levels, and that class 3 alleles seemed to be strongly associated with macroangiopathy in NIDDM subjects.24)
Mandrup-Poulsen et al.18) studied insulin gene polymorphism in 41 non-diabetic, Danish Caucacian patients with severe atherosclerosis defined by coronary arteriography. The frequency of the class 3 allele in those subjects was 0.30 compared with 0.12 in 21 non-diabetic control subjects.
We have found a very low frequency of a large (class 3) allele in both Korean diabetics and non-diabetics compared with the previous paper in Caucacians,7ŌĆō12) American blacks,13) and Pima Indiands.14) This difference in findings may be due to race difference and because of low incidence of atherosclerosis of Korea. Studies25ŌĆō27) in maturity onset diabetes in young (MODY) families have failed to identify an insulin allele that segregates with diabetes, but other families of varied racial background must be studied before insulin gene defects can be ruled out as a common cause of diabetes. Recently our diets have been westernized, and the incidence of diabetes mellitus (DM) and atherosclersos has increased. So in a futrue, we may guess raising genetic expression of DM by the environmental factor as diet, air and water pollution etc. Although low incidence of large allele was observed in the Korean poeple, we may use RFLP of the insulin gene region as a weak genetic marker. In the different point of view, at the first we checked RFLP of insulin gene in Korean acromegalics who were representatives of secondary DM, but they had no class 3 allele. This finding meant that expression of DM was concerned about factors other than DNA level. One possibility is that insulin gene biosynthesis is positively regulated by glucose concentration on translational level.28) Our task is further investigate to Korean characteristics of NIDDM and various secondary DM in genetic level and its relationship to disease entity.




 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Download Citation
Download Citation Print
Print





