INSTRODUCTION
Lasting and continuous competence is imperative as the field of medicine is constantly evolving. The need to investigate multiple populations in medical research has led to multicenter and even multinational trials. In multicenter trials, many challenges arise that are not present in single-institution studies, particularly with respect to clinical laboratory results. To overcome challenges associated with compiling and comparing results from different laboratories, standardization or harmonization of laboratory test results is needed. However, standardization and harmonization consume much time and resources, creating obstacles in multicenter trials [
1].
In multicenter trials, clinical laboratory testing can be performed in the laboratory of each participating site or in a central laboratory. In general, routine tests, such as complete blood cell counts, general chemistries, and urinalysis, are usually performed at the respective laboratory of each participating site; and special tests, such as drug concentration and genetic testing, are performed in a central laboratory. Because researchers may assume that various methods for measuring or evaluating an analyte produce the same results, they may not be aware of variability in results between methods, which could result from a lack of traceability. As all laboratories do not use the same analytic methods, measurement principles, calibrators, and reagents, test results may vary based on the laboratory, which makes comparison of test results from different laboratories difficult. In multicenter trials carried out to establish diagnostic or therapeutic guidelines or to aid in drug development, variance of clinical laboratory test results caused by the use of different measurement methods should be considered in evaluating results for optimal guidelines and efficient drug development. If these variances are not considered, the accuracy of the analysis may suffer and result in negative clinical, technical, financial, and regulatory consequences [
2].
In its 2015 survey, the College of American Pathologists (CAP), one of the largest external quality assessment organizations in the world, showed that the inter-assay coefficient of variation (CV) of total cholesterol was 3.3% across all methods, 3.9% for creatinine, and 3.2% for hemoglobin A1c [
3]. Those CVs were much lower than those of other analytes.
As international guidelines were established in hyperlipidemia and diabetes, standardization in measuring glucose and lipids were developed by using reference materials and methods [
1]. Standardization and harmonization are processes used to equalize results derived using different methods. Standardization can be accomplished by relating the result to a reference through a documented, unbroken chain of calibration. When such a reference is not available, harmonization is used to equivalize results utilizing a consensus approach, such as application of an agreed-upon method mean [
2]. However, accurate standardization rely on securing traceability from reagent manufacturers. Each laboratory in a trial must check the traceability of reagents in clinical laboratory tests before the trial begins. Although traceability may be confirmed, commutability of reference materials should also be considered. Without such standardization, harmonization, and traceability, accurate interpretation of trial results may be difficult. This study was conducted to evaluate the CV of laboratory results produced by various measuring methods and to determine whether mathematical data adjustment could achieve harmonization between the methods.
METHODS
Materials and methods
This study is part of the Cooperative Network Construction of a Nationwide Clinical Trial study [
4] to evaluate the characteristics of and treatment strategies in patients with hypertension in 37 Korean centers. Of these 37 centers, the laboratories of nine centers were investigated in this study. This study was approved by the Institutional Review Board of Cheil General Hospital & Women’s Healthcare Center (approval number: CGH-IRB-2013-33) and its associated centers, and written informed consent was obtained from all patients.
Basic data gathering
Nine laboratories (labeled A through I) as well as Green Cross Laboratories (GC Labs), the reference laboratory, were included in this study (
Fig. 1). Instruments, analytic methods, reagents, lot information of reagents, and traceability of calibrators were surveyed for the six test items, serum total cholesterol, high density lipoprotein cholesterol (HDL-C), low density lipoprotein cholesterol (LDL-C), triglycerides, creatinine, and glucose, in all 10 laboratories. The traceability chain is shown in
Fig. 2.
Manufacturing serum panel
A serum panel with 20 concentrations for each analyte was created for method comparison. Remaining patient samples at GC Labs were used to make the serum panels according to the Clinical & Laboratory Standards Institute EP9-A2 guideline. After 40 mL of the pooled serum samples were centrifuged at 3,000 rpm at 2°C to 8°C for 10 minutes, the supernatant was separated; and the supernatant was mixed for 10 hours under refrigerated conditions. Following filtration with a 0.22 μM filter (MF membrane, Sigma-Aldrich Co. LLC, St. Louis, MO, USA), 300 μL per tube were dispensed and kept in a deep freezer at –70°C before transportation to each participating laboratory under freezing conditions.
Measurement method
Each laboratory was asked to store samples in a deep freezer the day before measurement. All samples transported to each laboratory were analyzed in 14 days after being frozen. The analysis was done at the same time in both GC Labs and each laboratory to exclude bias due to pre-analytical conditions. Samples were thawed in a refrigerator for 1 hour and then were mixed for 30 minutes on a roller mixer. Each sample was analyzed in duplicate. The second measurement was done in reverse order compared to the first one. All measurements were done within 2 hours. Test results were reported in integers for serum total cholesterol, HDL-C, LDL-C, triglycerides, and glucose. Creatinine was reported to two decimal places.
Analytic method and traceability
Instruments used to measure the analytes in this study were as follows: modular analytics (Roche Diagnostics, Manheim, Germany) for GC Labs; Cobas 8000 c702 (Roche Diagnostics) for laboratories A, E, and G; TBA-2000FR (Toshiba Medical System Corporation, Tochigi, Japan) for laboratories B, D, H, and I; ADVIA1800 (Siemens Healthcare Diagnostics, Marburg, Germany) for laboratory C; and TBA 200FR NEO (Toshiba Medical System Corporation) for laboratory F (
Supplementary Table 1). Reagent and calibrator information for each analyte are described in
Supplementary Table 2. Ranges of CVs for the six analytes at all concentrations are shown in
Supplementary Table 3. The number of laboratories using the same calibrator and assigned values as the reference laboratory, GC Labs, was two for total cholesterol, one for LDL-C, two for triglycerides, two for glucose, and none for HDL-C and creatinine.
As seen in
Table 1, the analytic method for total cholesterol in all 10 laboratories was uniformly enzymatic. All methods were traceable with isotope dilution mass spectrometry or the Abell-Kendall method. For HDL-C and LDL-C measurements, direct methods were used (
Table 1). Direct methods do not include pre-analytical processes of ultracentrifugation, precipitation, and calculation steps, which make direct methods suitable for auto-analyzers. Every laboratory used the direct method using cationic detergent, and all laboratories used reagents traceable to the Centers for Disease Control and Prevention reference method (
Table 1) [
5-
7]. All laboratories used enzymatic methods for triglycerides. Five of the 10 laboratories used glycerol blank methods. For serum creatinine measurements, nine laboratories used the Jaffe method, one used an enzymatic method, and two did not use adjustment of pseudo-creatinine chromogen [
8,
9]. All used the hexokinase method to measure glucose. Traceability was ensured with reference materials for glucose.
Statistics
For statistical analysis, EP evaluator release 11 (Data Innovations LLC, South Burlington, VT, USA) and Excel 2000 (Microsoft Corp., Redmond, WA, USA) were used. CV was calculated for central and each laboratory variation from each sample. The interassay CVs for each analyte was calculated by averaging the observed CVs over all 20 samples. We aimed to find the line of bestfit using by Deming regression equation, which was defined y-axis as a reference standard and x-axis as each laboratory result (y = β0 + β1 × X). To evaluate the effect of harmonization using Deming regression analysis, interassay CVs before and after harmonization were calculated. Differences in the proportion of diseases before and after adjustment for lab harmonization were analyzed by a proportion test. For box plot and strip charts, R general-public-license version (R Foundation, Vienna, Austria) was used.
DISCUSSION
This study showed that result variation caused by different analytical methods can be reduced by harmonization. Harmonization may become a prerequisite in multicenter trials. The compatibility of data generated by multiple laboratories is not guaranteed due to different methods, reagents, calibrators, etc., used; and management of data from multiple sites is difficult and requires more effort for statistical analysis than what is needed in single-center studies [
10].
When clinical laboratory tests may be performed by the laboratory of each participating site, more discussion and consideration in the planning stage of research should be given to whether a single, central clinical laboratory should be used instead of the laboratories at each center. If multiple laboratories will be analyzing samples, researchers should explore how to adjust or compare data prior to initiating the study.
This study was carried out to establish a method of postanalytical harmonization using data from various laboratories. The clinically acceptable total error, including precision and bias, is reported in international guidelines, the literature, and reports from external quality assessment organizations [
11-
13]. For example, in the CAP survey (external quality assessment), interassay CV among all methods for total cholesterol is 3.0% to 3.4%, 3.9% to 15.5% for creatinine, and 3.2% to 9.2% for glucose, which are rather good [
3]. Accuracy of the analytic method used in clinical laboratories is established by a standardization process using the results from patient samples, the hierarchy structure, and traceability of the analytical measurement system (
Fig. 1) [
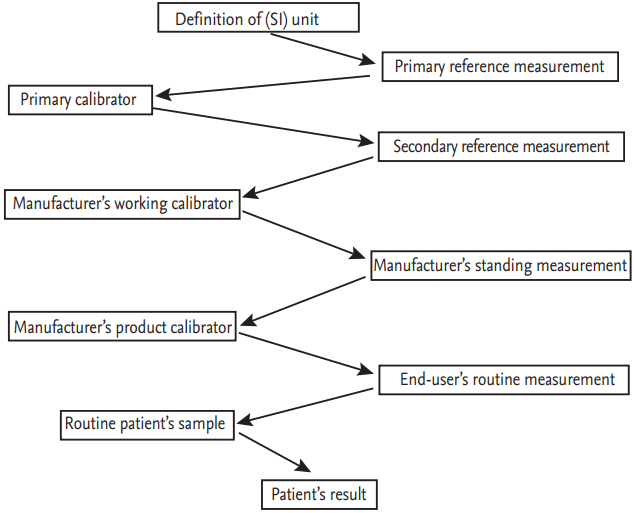
14]. This association ensure traceability of analytical methods. Traceability applies a primary reference material with proper accuracy and precision, by cascades of reference material, to the calibrator by the manufacturer, and finally to the patient samples (
Fig. 2). The interassay CV of test results from different laboratories is small for an analytical method with traceability that uses the same calibrator adjusted with the same primary reference material. This is standardization. All reagents used in this study have traceability. Thus, when compared with external quality assessment scheme results, all analytes had better or similar results except triglycerides (
Supplementary Table 3).
Our study results are also concerning. Although interassay CVs were within acceptable limits according to external quality assessment standards and comparison studies revealed good correlation coefficients, the results of some of the participating laboratories should be evaluated for adjustment against international guidelines like those from the National Cholesterol Education Program. International guidelines propose strict criteria, and not all our results could meet those requirements. In studies where the analysis of the crude data could influence the study’s conclusion, failure to adjust the results could lead to challenges to the integrity of the study. Traceability is particularly important in multicenter trials where the different laboratories utilize various testing methods. However, even where the methods are traceable, differences between the various methods should not be ignored because commutability of reference material, application of the calibrated assigned value, and method compensation, such as the creatinine determination method, could be problematic [
2]. Thus, traceability of methods coupled with studies comparing the results from different laboratories during the planning stages of multicenter trials would guide adjustments of results so that analysis of results from multiple sites would be accurate. As inter-assay CVs of the laboratories using the same calibrator and assigned value showed lower variance than the CVs of those that did not, traceability should be checked first when selecting analytical methods in multicenter trials.
Different methods impact CV differently. As the analytic method for triglycerides is affected by the use of a glycerol blank, the method use by each laboratory should be surveyed and considered in the interpretation of test results. Measuring triglyceride concentration without a glycerol blank has a high positive correlation with free glycerol concentration, which was also shown by regression analysis [
15], so that methods with or without glycerol blank could be compared by regression analysis. In the current study, when results from the nine participating laboratories were adjusted based on the method used in the central laboratory, interassay CVs were then greatly improved. Compared with the 2015 CAP survey, interassay CVs of the participating laboratories were high. The reason may be that, in our group, 50% used a method with a glycerol blank, whereas, in the CAP group, only 10% used a method with a glycerol blank [
3]. Data adjustment would greatly improve accuracy when analytic methods of participating laboratories demonstrate systemic bias.
In creatinine analysis, the Jaffe method and enzymatic method can be used. The differences between the analytic methods depend on avoiding interference of pseudo-creatinine chromogen, such as proteins, antibiotics, and ketones. Different measurements using the Jaffe method may yield significant analytic errors depending on whether compensation was made for those chromogens [
8] because creatinine is a very low-concentration analyte. For such reasons, interassay CVs before and after adjustment were higher for creatinine than for other analytes. After adjustment, the interassay CV for creatinine decreased dramatically, and the correlation coefficient was good.
To measure glucose, all laboratories used the hexokinase method. As traceability was secured with reference methods and materials, the mean CV for glucose was excellent at 1.7% (0.7% to 3.4%).
Using a single, assigned analytic method in a central laboratory would be best in multicenter trials. However, because of the logistical challenges to sample transportation and the need for timely analysis, analyzing samples in the laboratories of the participating center where the sample was taken is how many multicenter trials currently manage the analysis. In this situation, data harmonization is an option. Laboratory data harmonization is especially needed when reference methods or materials are absent or when reference material is non-commutable. As described above, calibrator values and method characteristics can influence results and, thus, analysis of data. For detecting these possible obstacles to accurate analysis and deciding on harmonization, pre-study surveys of the participating laboratories should be considered. In multicenter trials, direct comparison without data adjustment is best practice provided no variance in results existed; but, in older studies, traceability was seldom addressed, complicating comparisons between data sets. This study confirms the need to compare assays and methods used in obtaining older data and to adjust data if needed. In the future, if a central laboratory could compare its results with those of a reference laboratory, such as Cholesterol Reference Method Laboratory Network, and calculate bias, the central and participating laboratories could coordinate using a hierarchical structure permitting the central laboratory to communicate the reference material obtained to the participating laboratories and creating consistency in measurement of analytes across the participating laboratories.
The major limitation of our study is no external validation of this model in clinic. Although the prevalence of diseases such as dyslipidemia and chronic kidney disease changed significantly after application of our proposed lab harmonization method, it is still strongly required to test whether this regression reduces the interlaboratory gap. Further studies are needed.
Perspective
This study will encourage researchers to focus on traceability and improve the quality of study results through harmonization. For analytes with available reference materials and methods, standardization would be the best approach; and harmonization should be applied to the analytes for which reference materials and reference methods are not available or cannot be developed. Additional studies on harmonization of reference ranges may broaden the scope of the interpretation of clinical laboratory results [
16].







 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Download Citation
Download Citation Supplement 1
Supplement 1 Print
Print



